In-vitro detection method for quickly screening exon 5-8 mutation of P53 gene and application
A detection method, an in vitro detection technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem of not being able to determine the position of mutations in DNA fragments, and facilitate high-throughput processing , high detection sensitivity and low dosage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] An in vitro detection method for rapid screening of P53 gene exon 5-8 mutations, comprising the following steps:
[0032] (1) Extraction of Dried Blood Spot DNA
[0033] The total DNA extraction was carried out according to the instructions of the Magnetic Bead Method Dried Blood Spot Genomic DNA Extraction Kit (Tiangen), as follows:
[0034] 1. Take a new 96-well deep-well plate, and add three 3*3mm dried blood spot samples to each well. Add 200 µL of buffer GA to a 96 deep-well plate.
[0035] 2. Add 20 μL of proteinase K solution, vortex for 10 seconds to mix well, put it into a constant temperature oscillator preheated to 56 degrees, and lyse at 900 rpm for 30 minutes.
[0036] 3. During the sample lysis in the previous step, add 10 μL magnetic bead suspension G, 200 μL buffer GB and 200 μL absolute ethanol to a new 96-well deep-well plate, and mix by whipping or shaking for 10 seconds.
[0037]4. After the sample is lysed, briefly centrifuge the deep-well plate ...
Embodiment 2
[0056] Embodiment 2: clinical sample detection
[0057] The method of Example 1 was used to detect exons 5-8 of the P53 gene in the blood of 200 cases of physical examination population, and the mutation of the P53 gene in the blood DNA of 3 cases of subjects could be successfully detected, and then the PCR product was analyzed by using the next generation sequencing method. After detection and comparison with the human genome sequence, it was confirmed that there were indeed changes in the DNA sequence of these three samples.
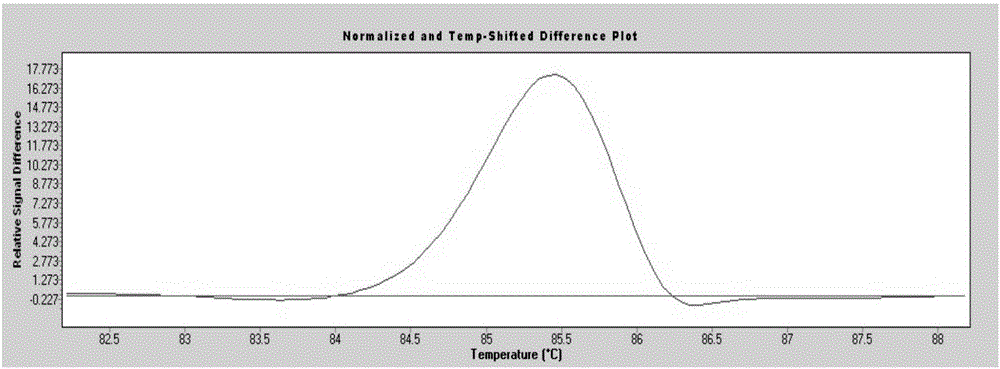
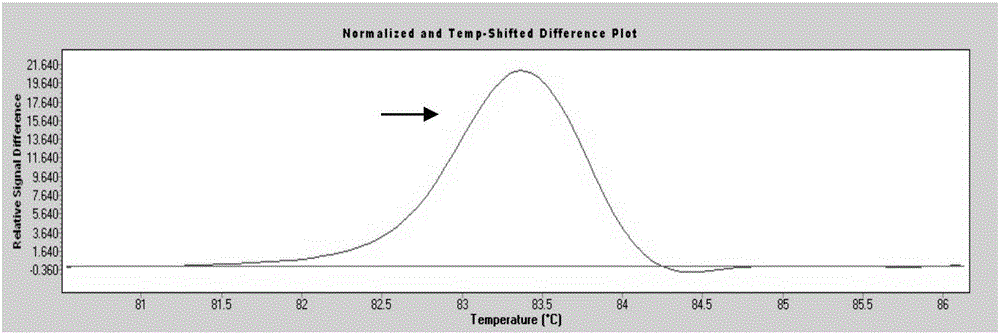
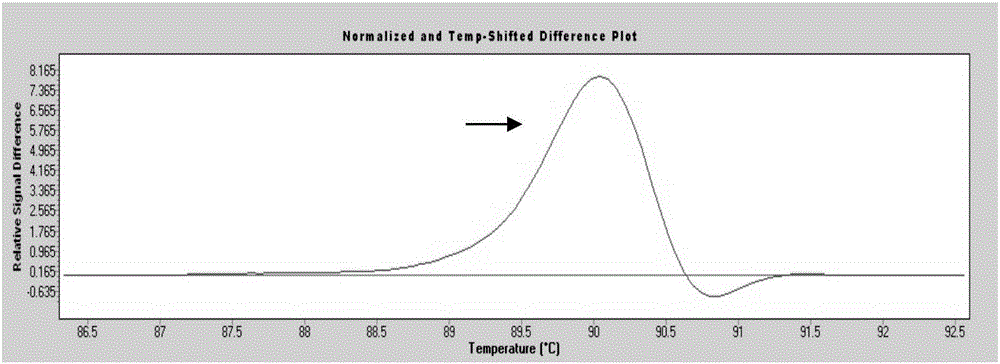
[0058] Among the 200 blood samples from the physical examination population, 3 cases (No. 68, No. 105, and No. 160) had mutations (separate peaks appeared), and No. 68 sample P53 gene exon 7 was suspected to have a nucleic acid sequence change ( figure 1 ); No. 105 sample P53 gene exon 6 is suspected to have a nucleic acid sequence change ( figure 2 ); Nucleic acid sequence change in exon 5 of P53 gene in sample No. 160 was suspected ( image 3 ). ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com