Gene editing of pcsk9
An editing and encoding technology, applied in genetic engineering, metabolic diseases, extracellular fluid diseases, etc., can solve problems such as genome instability and off-target effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0596] Example 1: Guide-guide nucleotide sequences-programmable DNA-binding protein domains, deaminase domains and base editors
[0597] Non-limiting examples of suitable leader nucleotide sequences-programmable DNA binding protein domains are provided. The present disclosure provides Cas9 variants, such as Cas9 proteins from one or more organisms, which may comprise one or more mutations (eg, to generate dCas9 or a Cas9 nickase). In some embodiments, one or more of the amino acid residues of the Cas9 protein (identified with an asterisk below) can be mutated. In some embodiments, the D10 and / or H840 residues of the amino acid sequence provided in SEQ ID NO: 1, or the corresponding mutations in any of the amino acid sequences provided in SEQ ID NO: 11-260, are mutated. In some embodiments, the D10 residue of the amino acid sequence provided in SEQ ID NO: 1, or the corresponding mutation in any of the amino acid sequences provided in SEQ ID NO: 11-260, is mutated to any resi...
Embodiment 2
[0689] Example 2: Methods for modifying PCSK9 and other liver proteins to improve circulating cholesterol and lipid levels CRISPR / Cas9 genome / base editing method
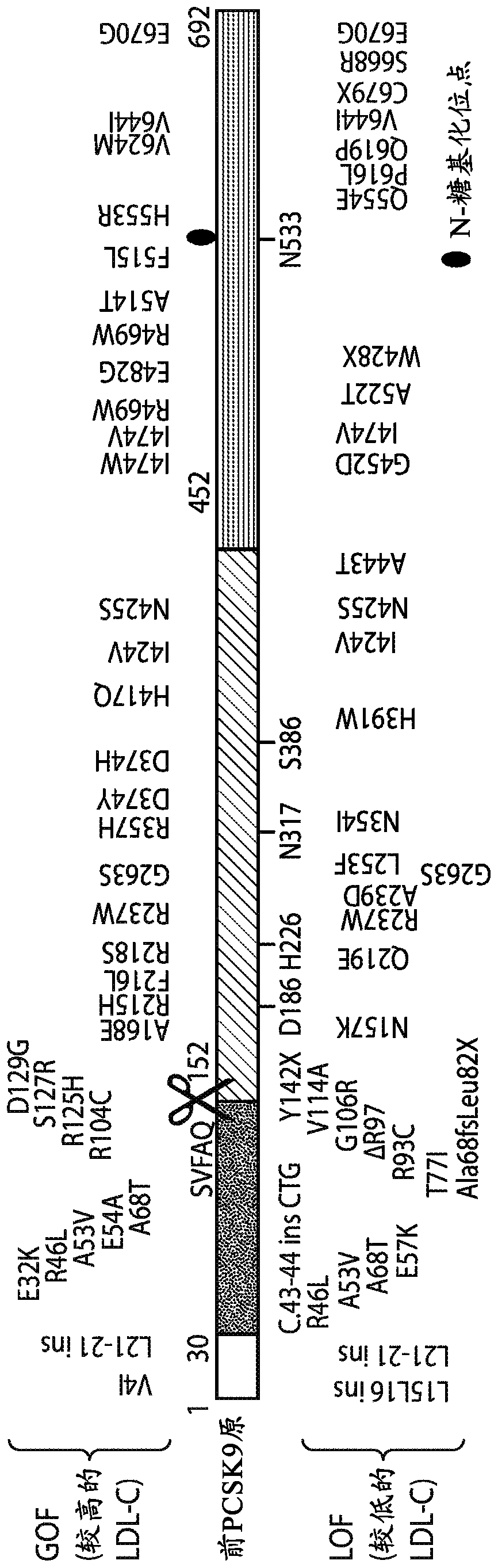
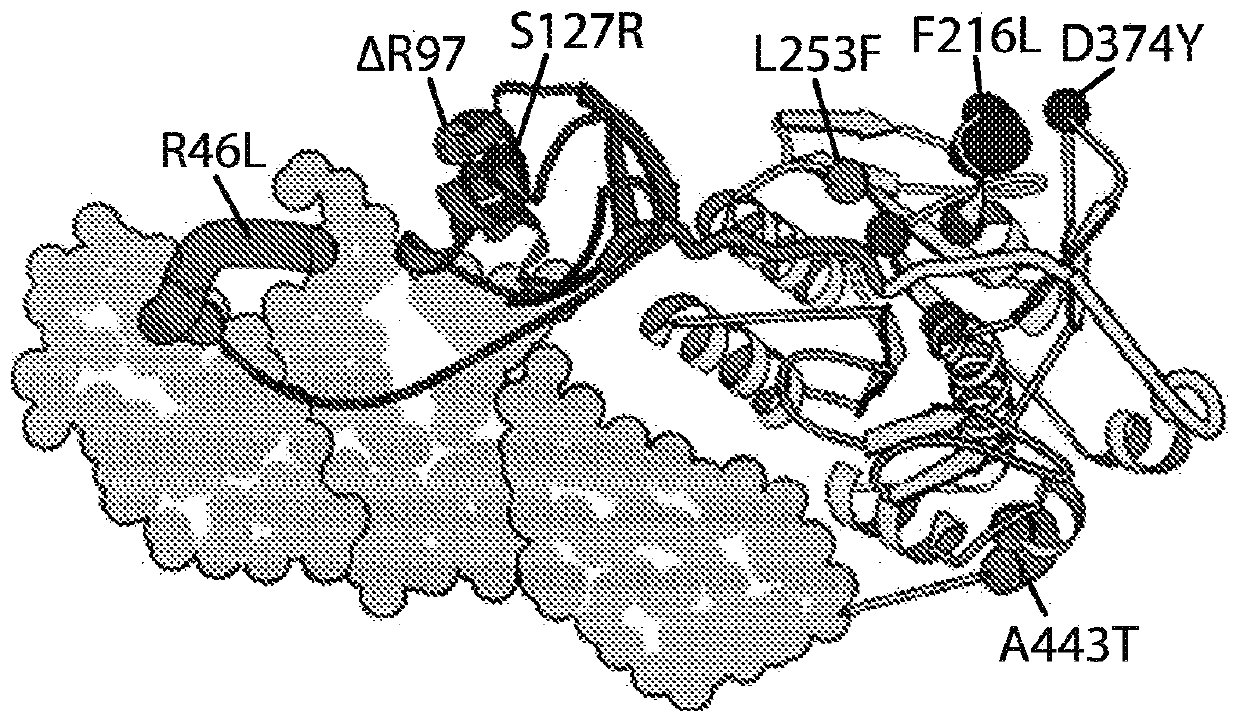
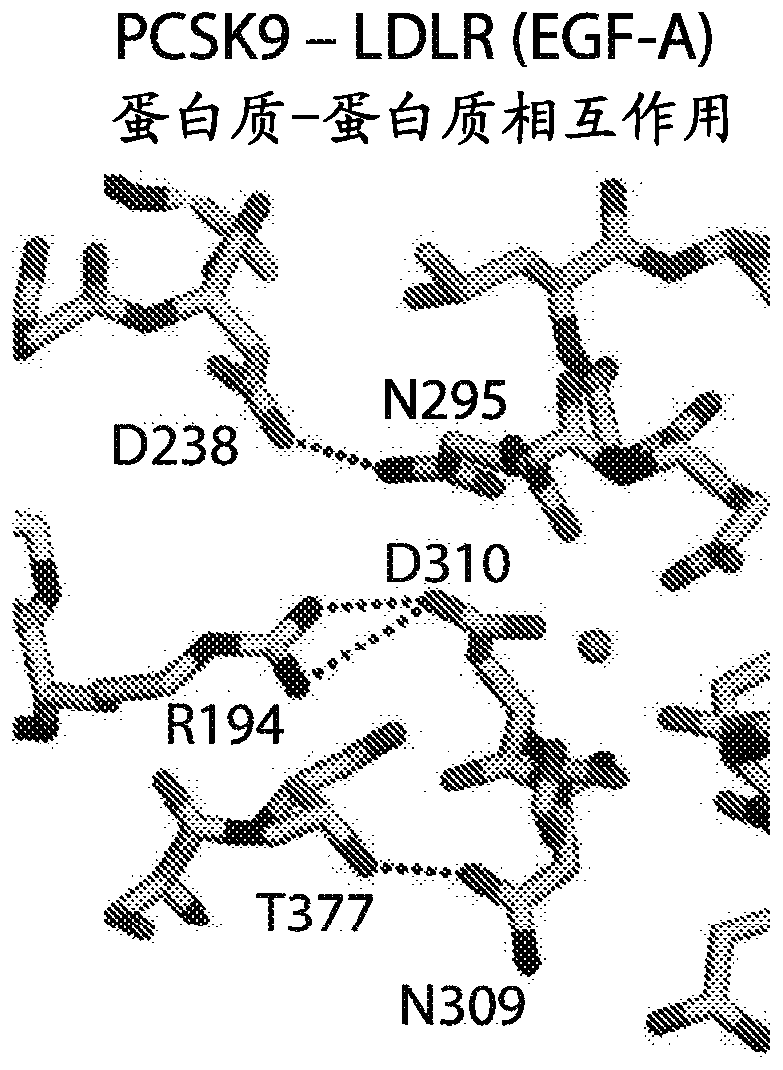
[0690] Approximately 70% of circulating cholesterol is transported within low-density lipoproteins (LDL), which are cleared in the liver by LDL receptor (LDL-R)-mediated endocytosis, along with endogenous Additional consequences of downregulation of the cholesterol biosynthetic pathway. PCSK9 is a secreted, globular serine protease capable of proteolytic autoprocessing of its N-terminal prodomain into a potent endogenous inhibitor that permanently blocks its catalytic site ( Figures 1A to 1C ). A list of agents used to block PCSK9 function can be found in Table 12. Mature PCSK9 exits through the secretory pathway and functions as a protein-binding linker in clathrin-coated vesicles to bridge the pH-dependent interaction with the LDL receptor during endocytosis of LDL particles, which prevents LDL receptor re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com