Method for efficiently synthesizing PHBs by knocking out LPS synthesis gene cluster
A gene cluster and gene technology, applied in the fields of genetic engineering and fermentation engineering, can solve the problems that PHB cannot meet the needs of industrial production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] The construction of embodiment 1 engineering bacteria WJW00
[0045] The engineering bacteria WJW00 has been published in the SCI paper "Construction and Characterization of an Escherichia coli Mutant Producing Kdo 2 -Lipid A" (disclosure date: March 13, 2014). The specific construction process is as follows:
[0046] (1) Obtaining of gmhD (also known as rfaD) gene knockout fragment
[0047] The gmhD gene knockout fragment is obtained by chemical total synthesis or PCR step-by-step amplification, and its two ends are the upstream and downstream homology arms of the gmhD gene, and the middle is a kan fragment with an FRT site. The nucleotide sequence of the gmhD gene knockout fragment is shown in SEQ ID NO.2. The gmhD gene knockout fragment was cloned into the plasmid pBlueScriptIISK(+) to obtain the recombinant plasmid pBlueScript IISK(+)-gmhDU-Fkan-gmhDD. Using the plasmid as a template, the knockout fragment gmhDU-Fkan-gmhDD can be amplified.
[0048] (2) Prepara...
Embodiment 2
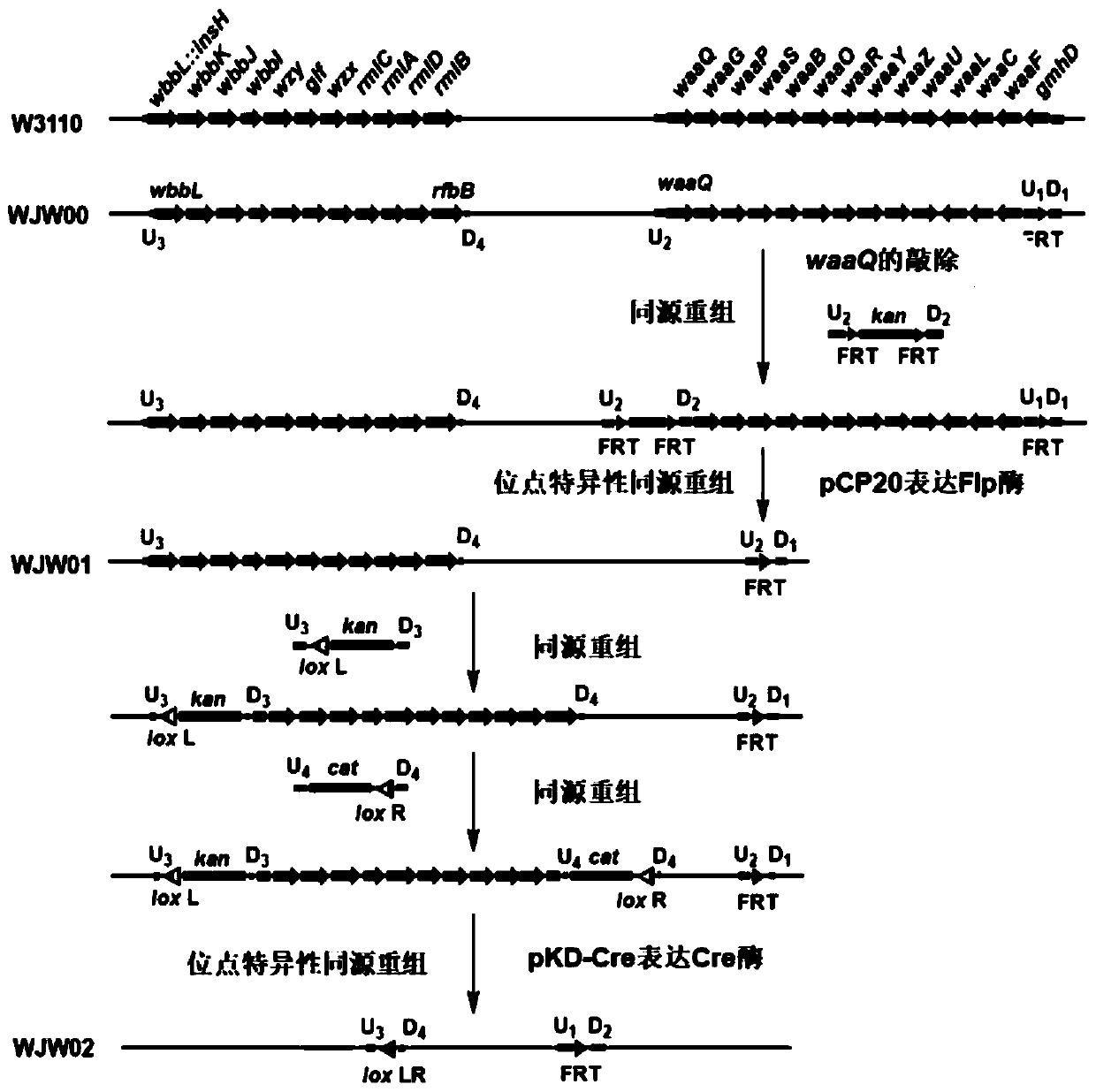
[0054] Example 2 Construction of LPS streamlined strain WJW01
[0055] Lipopolysaccharide (LPS) includes three parts: lipid A, core sugar and O-antigen, among which lipid A is a highly conserved region, while core sugar and O-antigen are composed of different sugar molecules. Among them, the core sugar synthesis gene cluster contains 14 genes, namely waaQ, waaG, waaP, waaS, waaB, waaO, waaR, waaY, waaZ, waaU, waaL, waaC, waaF, gmdD (also known as rfaD), the NCBI of its sequence The accession numbers are "BAE77660.1", "BAE77661.1", "BAE77662.1", "BAE77663.1", "BAE77664.1", "BAE77665.1", "BAE77666.1", "BAE77667.1 ", "BAE77668.1", "BAE77669.1", "BAE77670.1", "BAE77671.1", "BAE77672.1", "BAE77673.1".
[0056] On the basis of the strain WJW00 obtained in Example 1, the remaining 13 genes in the core sugar gene cluster gmdD(rfaD)-waaFC-waaQGPSBORYZUL except the gmhD gene were knocked out.
[0057] (1) With reference to the knockout process of the gmhD gene in Example 1, the waaQ g...
Embodiment 3
[0071] Example 3 Construction of LPS streamlined strain WJW02
[0072] The O-antigen synthesis gene cluster rmlBDACX-glf-wbbHIJKL contains 11 genes, namely wbbL::IS5, wbbK, wbbJ, wbbI, wzy, glf, wzx, rmlC, rmlA, rmlD, rmlB, and their sequence accession numbers are " BAA15873.1", "BAA15874.1", "BAA15875.1", "BAA15876.1", "BAA15877.1", "BAA15878.1", "BAA15879.1", "BAA15880.1", "BAA15881. 1", "BAA15882.1", "BAA15883.1", "BAA15884.1". WJW02 was obtained after further knocking out the O-antigen synthesis gene cluster on the basis of WJW01. The specific operation is as follows:
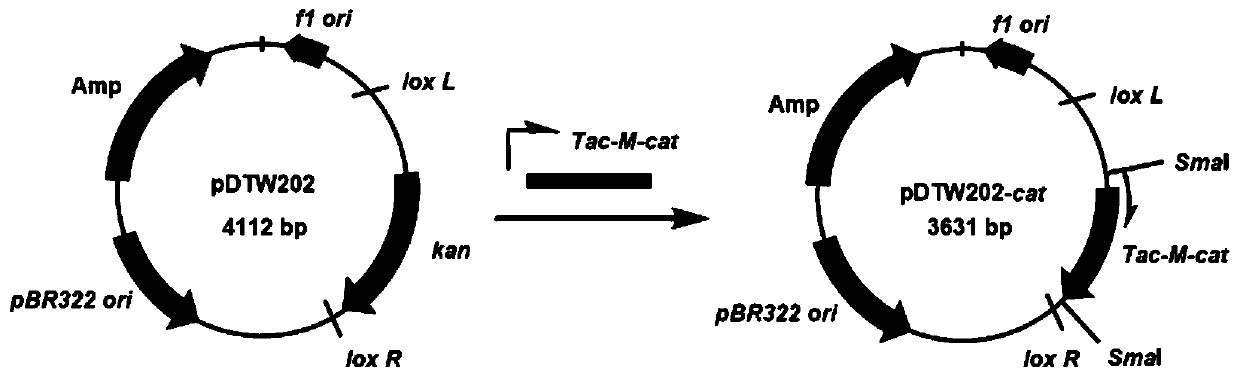
[0073] (1) Construction of tool plasmid pDTW202-cat
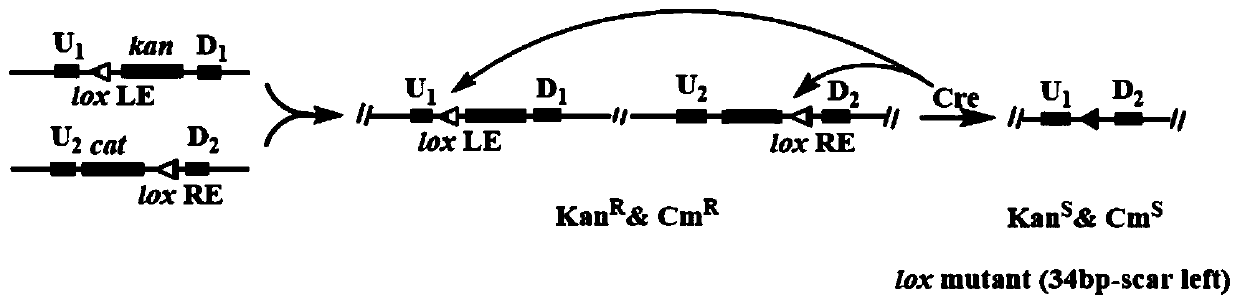
[0074] The knockout of O-antigen gene cluster wbbL-rmlB adopts lox LR site-specific recombination method. Therefore, when constructing the Escherichia coli genome streamlining system, it is necessary to construct a tool plasmid pDTW202-cat. The construction process of the plasmid is specifically as follows:
[0075] 1) Using the plasmid pDTW109 (se...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com