Method and device for detecting somatic variation in samples based on single-sample next-generation sequencing
A next-generation sequencing, single-sample technology, applied in the field of bioinformatics, can solve the problems of inability to accurately find somatic cell variation, lack of white blood cells, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
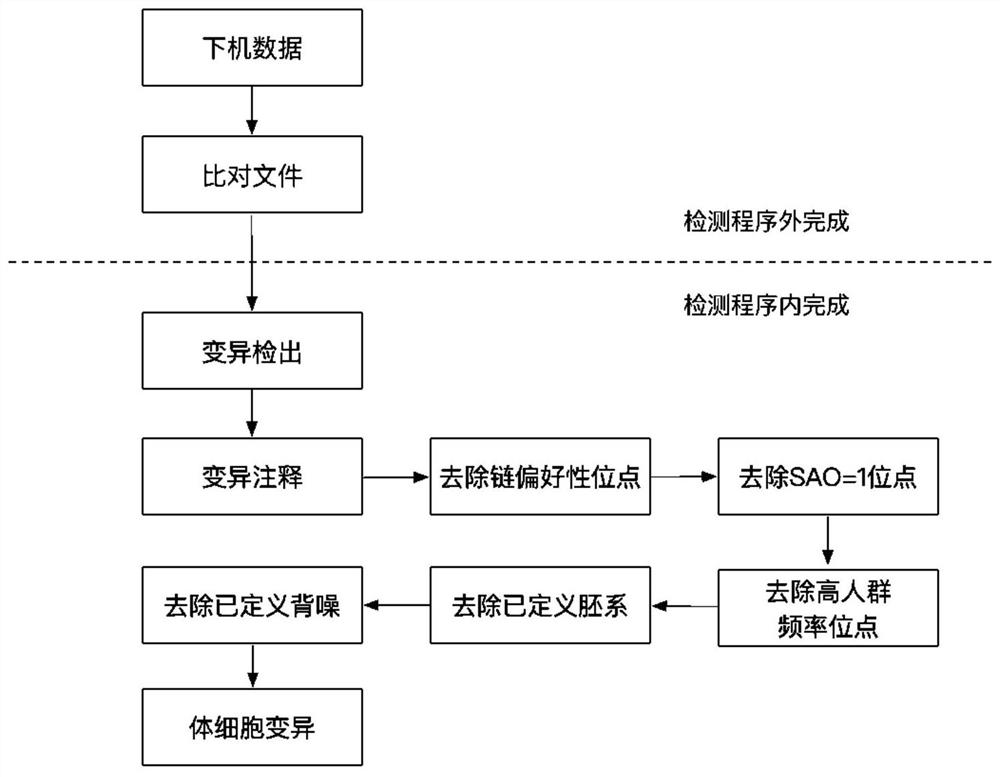
[0025] According to a typical embodiment of the present invention, the method further includes the step of annotating the detected variation, preferably, the annotation includes annotating amino acid changes, 1000 genome population frequency, ExAC according to the genomic position of the variation and the variation base Population frequency and information recorded in the dbSNP database, etc. Annotate public database information on all variants so that variants can be filtered using the public database information on the annotation. In one embodiment of the present invention, variant filtering includes: removing strand-preferred sites, preferably, strand-preferential sites refer to variants that support mutation reads in the positive or negative strands to be 0, so as to remove mutations due to amplification False positive variants due to bias.
[0026] According to a typical embodiment of the present invention, variant filtering further includes: removing variants annotated ...
Embodiment 1
[0048] In this embodiment, the specific operations are mainly divided into two parts, the first part is the part completed outside the detection program, and the second part is the part completed in the detection program.
[0049] In this embodiment, the sample to be tested is a single-sample lung cancer pathological sample.
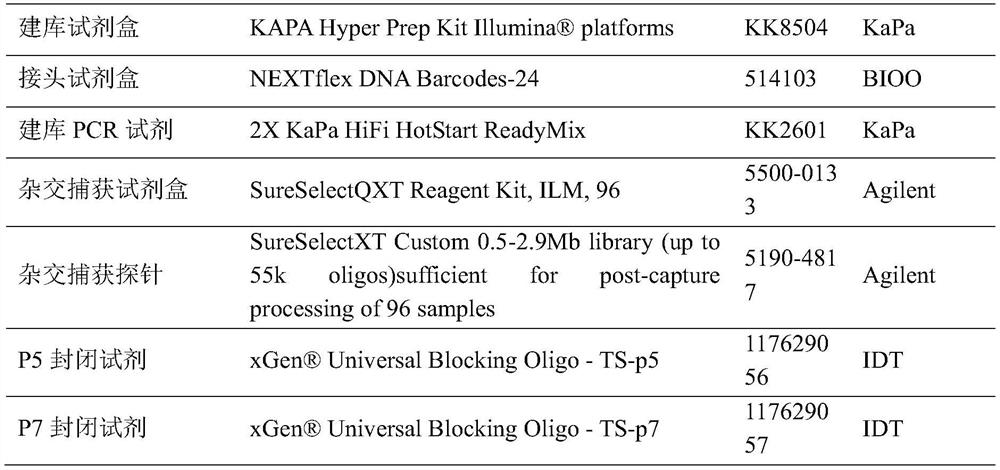
[0050] In the embodiment of the present invention, the main reagent supplies are commercially available, and the information is as follows in Table 1:
[0051] Table 1
[0052]
[0053]
[0054] The specific operation steps are as follows:
[0055] 1. The sample was pretreated and DNA was extracted, and quantified with a fluorescence quantitative meter (Qubit) at a concentration of 3.8 ng / μl and a volume of 130 μl; the sample was fragmented with a sonicator (Covaris) to make the DNA fragment size within 200 μl. Between ~400bp, and then use agarose gel electrophoresis to check whether the fragment size meets the requirements.
[0056] 2. The frag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com