PCR primer set for HLA gene, and sequencing method using same
A technology of HLA-C and HLA-DRB1, which is applied in the field of efficient and highly uniform sequencing, can solve the problems of difficult to detect alleles, reduce the uniformity of amplification and collection efficiency, and prevent non-specific amplification and reduce HLA Type cost, effect of reducing wrong determination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
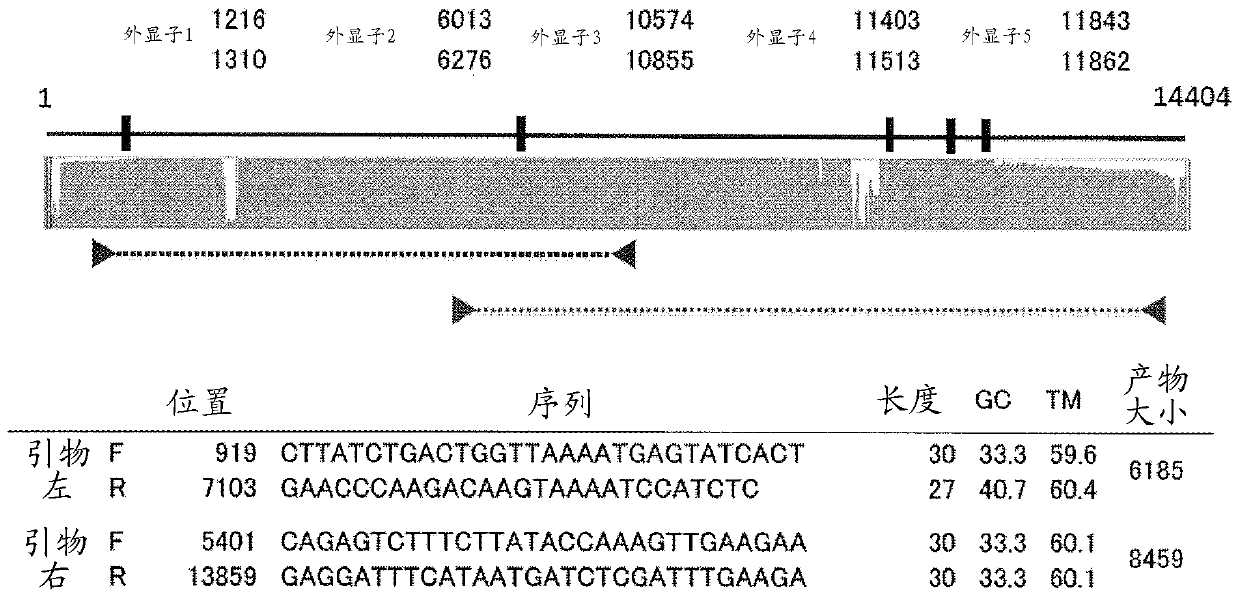
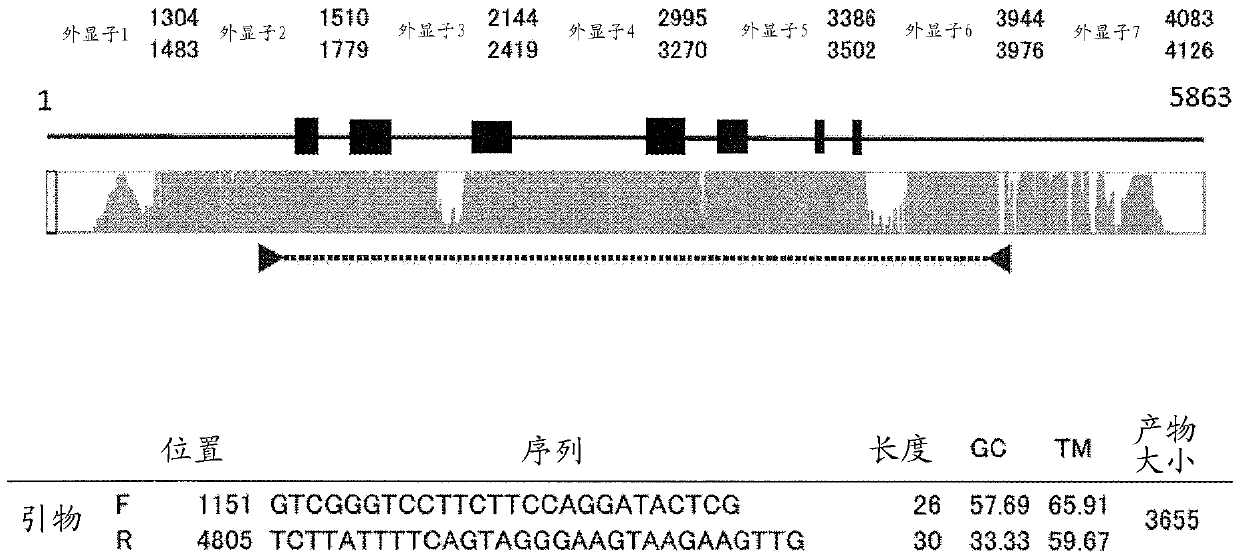
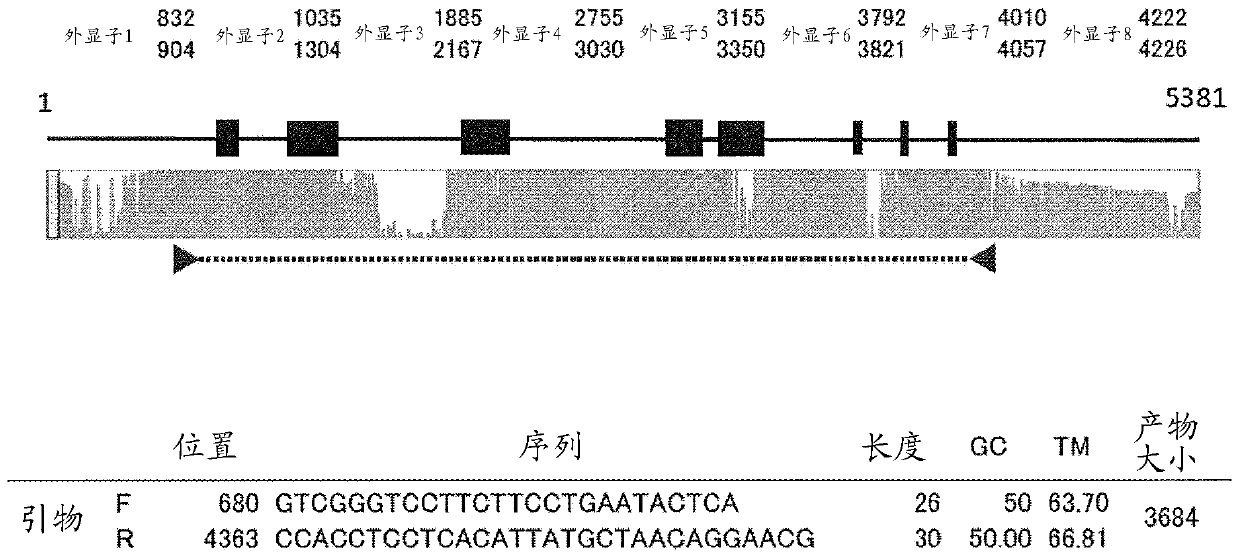
[0089] Embodiment 1 The design of the multiplex PCR primer set of classical HLA gene
[0090] A procedure for designing primers for multiplex PCR of the six classical HLA genes (HLA-A, -B, -C, -DRB1, -DQB1, -DPB1) is described. Bowtie2 (version 4.1.2) was used to map all reads obtained by the next generation sequencer.
[0091] I. Assembly of full-length genes from sequencing results of HLA genes
[0092] 1. Sequencing of HLA genes by conventional methods
[0093] Using an existing primer set [Hosomichi, K. et al. (2013) Phase-defined complete sequencing of the HLA genes by next-generation sequencing. BMC Genomics, 14:355.], 768 samples (unreleased samples) were Multiplex PCR with 2 panels (3 genes per panel) (see Table 2). For DRB1, primers with modified sequences were added. For the experimental conditions, refer to Table 3. The obtained amplification products were sequenced by MiSeq.
[0094]
[0095]
[0096] 2. Mapping to HLA dictionary
[0097]For the ...
Embodiment 2
[0174] Example 2 Multiplex PCR and NGS sequencing
[0175] I. Long PCR
[0176] 1. A total of 384 DNA samples were each subjected to multiplex long PCR targeting 6 genes of HLA-A, -B, -C, -DRB1, -DQB1, -DPB1. Amplifications were performed in the corresponding two pools below.
[0177] Set 1: HLA-A, -C, -DPB1(FF, FR), -DRB1(FF, FR)
[0178] Set 2: HLA-B, -DQB1, -DPB1(RF, RR), -DRB1(RF, RR)
[0179] Primer sequences and final concentrations are shown in Table 3. Prepare the following for PCR.
[0180] DNA 25ng
[0181] 0.25 μL of Tks Gflex TM DNA polymerase
[0182] 6.25 μL 2 × Gflex TM PCR buffer
[0183] After preparation to a final volume of 12.5 μL, apply Cool Start TM method (TaKaRa Inc.), and PCR reactions were performed under the temperature conditions and cycle numbers shown in Table 5. As a PCR device, use GeneAmp R PCR System 9700 (Thermo Fisher Scientific inc., Waltham, MA, USA).
[0184] 2. By Agencourt R AMPure R Amplified products were purifie...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com