Interactive parameterized three-dimensional DNA conformation simulation method for secondary school teaching
A simulation method and interactive technology, applied in the field of computer simulation, can solve problems such as the inability to give the geometric structure of DNA, the inability to clearly present the microscopic geometric structure of DNA molecules, and low user participation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0085] The present invention will be further described below in conjunction with the accompanying drawings and embodiments.
[0086] like figure 1 As shown, the present embodiment provides a method for interactive parametric simulation of three-dimensional DNA conformation for middle school teaching, based on the OpenGL standard graphics interface for simulation, including the following steps:
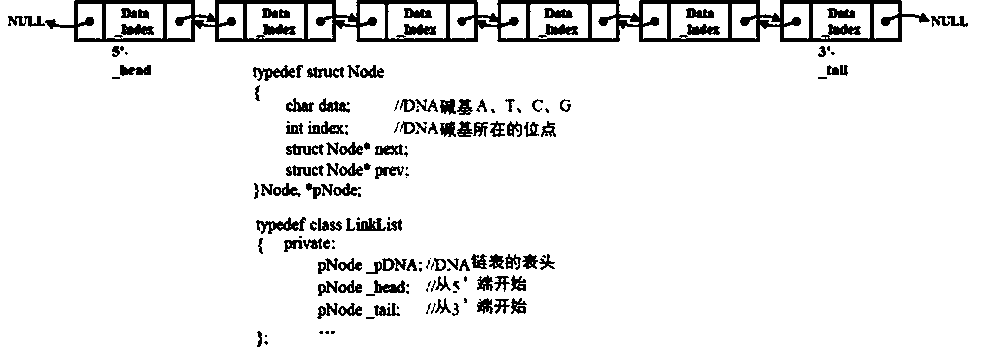
[0087] Step S1: In the VR / AR environment, according to the given error rate, the reverse complementary parallel primary structure of the DNA molecule is constructed in the form of orthographic projection, and the errors of the primary structure of the DNA molecule are corrected interactively;
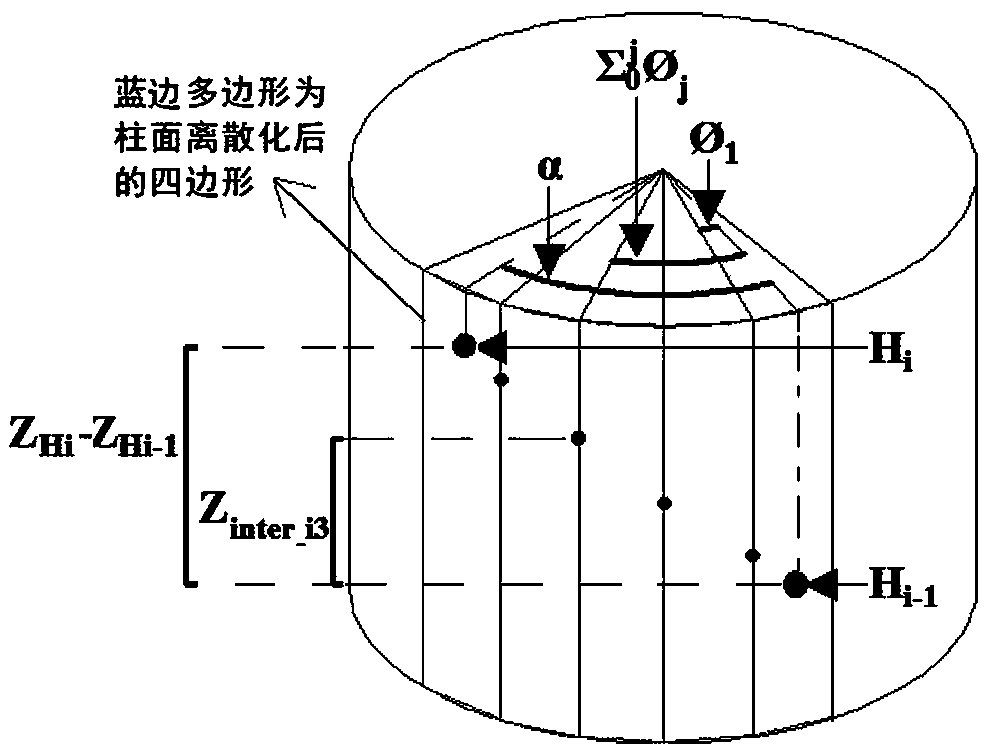
[0088] Step S2: In the VR / AR environment, using the primary structure established in step S1 as a reference, interactively simulate the dynamic generation process of DNA reverse double helix strands in three-dimensional space, and realize different forms of DNA secondary structures by adjusting ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com