Detection method of transformed DNA recovery rate and primers
A recovery rate and primer pair technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of lack of mutual confirmation of different detections
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Transformation of embodiment 1-DNA
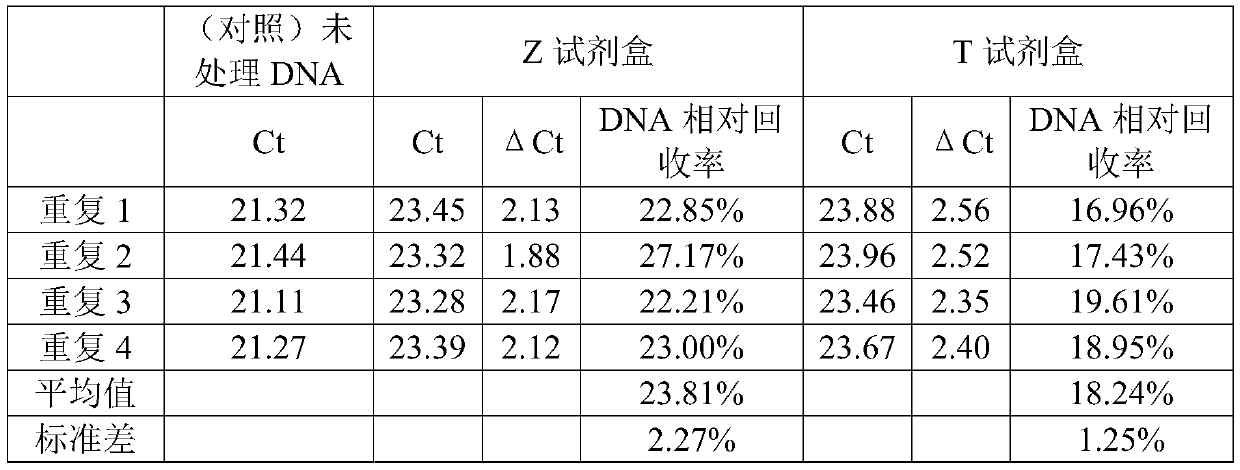
[0061] In this example, the Z kit and the T kit were used to transform DNA. The fragmented reference genome NA12878 DNA was used as the detection DNA, and each kit was repeated 4 times for detection.
[0062] 1) Configure CT conversion reagents according to the instructions of each kit.
[0063]2) Adjust the concentration of fragmented NA12878 to 1 ng / μL, take 20 μL and add 130 μL CT conversion reagent, and repeat the detection 4 times for each kit. Keep about 20 μL of untransformed DNA as a control.
[0064] 3) Run the CT conversion temperature control program according to the instructions of each kit. The Z kit is 98°C for 8 minutes, 54°C for 1 hour, and stored at 4°C. The T kit is 98°C for 10 minutes, 64°C for 2 hours and 30 minutes, and stored at 4°C.
[0065] 4) Purify the post-conversion DNA adsorption column according to the kit instructions, including DNA binding, desulfonation reaction, washing and elution. Finally, 20...
Embodiment 2-C
[0066] Example 2-CFF qPCR detection
[0067] In this example, CFF primers were used to perform qPCR on the transformed DNA to obtain the Ct value.
[0068] 1) Dilute the CFF forward and reverse primers to 4 μM, respectively, take the same volume and mix to form a primer mixture, and configure the qPCR reaction mixture as follows: Kapa SYBR Fast qPCR MIX 5 μL, CFF primer mixture 1 μL, and control or transformed DNA 4 μL.
[0069] CFF forward primer: TAAGAGTAATAATGGATGGATGATG
[0070] CFF reverse primer: CCTCCCATCTCCCTTCC
[0071] 2) Vortex and briefly centrifuge.
[0072] 3) Put the PCR tube into the fluorescent quantitative PCR instrument (BioRad CFX connect Real-Time PCR Detection System), and run the following program:
[0073] Pre-denaturation: 95°C for 5 minutes;
[0074] 40 cycles of signal collection: 95°C for 30 seconds; 54°C for 60 seconds (fluorescence signal collection).
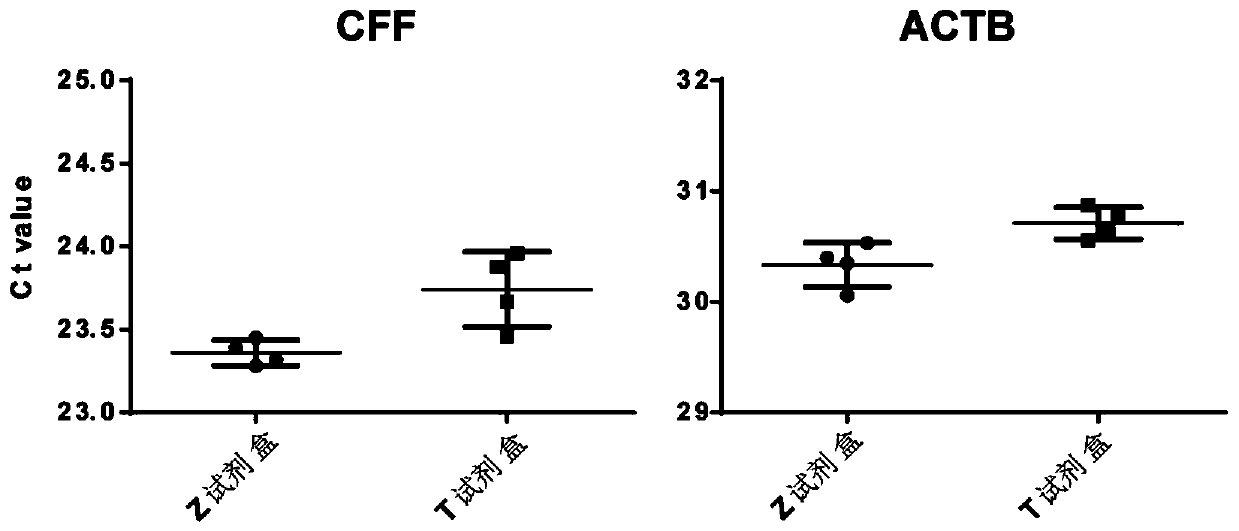
[0075] The results are shown in Table 1 and figure 2 shown. Statistical CFF primer dete...
Embodiment 3-A
[0080] Example 3-ACTB qPCR detection
[0081] In this example, qPCR was performed on the transformed DNA using ACTB primers to obtain the Ct value.
[0082] 1) Dilute the ACTB forward and reverse primers to 10 μM respectively, take the same volume and mix them to form a primer mixture, and dilute the ACTB probe to 4 μM.
[0083] 2) Configure the qPCR reaction as follows: NEB Luna PCR master mix 10 μL, ACTB primer mix 2 μL, ACTB probe 1 μL and transformed DNA 7 μL.
[0084] ACTB forward primer: GTGATGGAGGAGGTTTAGTAAGTT
[0085] ACTB reverse primer: CCAATAAAACCTACTCCCTCCCTAA
[0086] ACTB probe:
[0087] / 5TAMRA / ACCACCACCCAACACACAATAACAAACACA / 3IABkRQ /
[0088] 3) Put the PCR tube into the fluorescent quantitative PCR instrument (ABI 7500 Real-Time PCR System), and run the following program:
[0089] Pre-denaturation: 95°C for 5 minutes;
[0090] 40 cycles of signal collection: 95°C for 15 seconds; 58°C for 60 seconds (fluorescence signal collection).
[0091] The results ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com