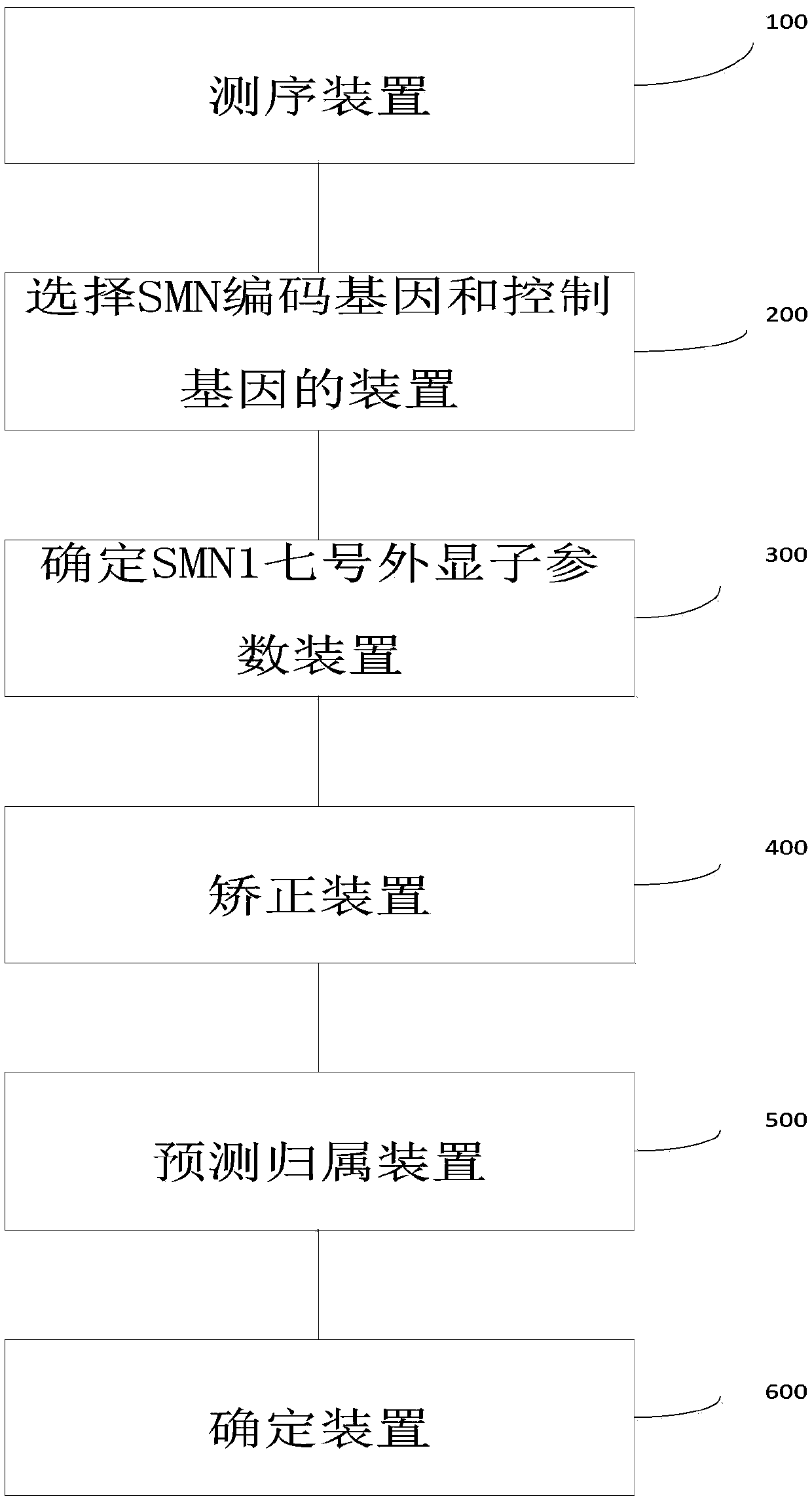
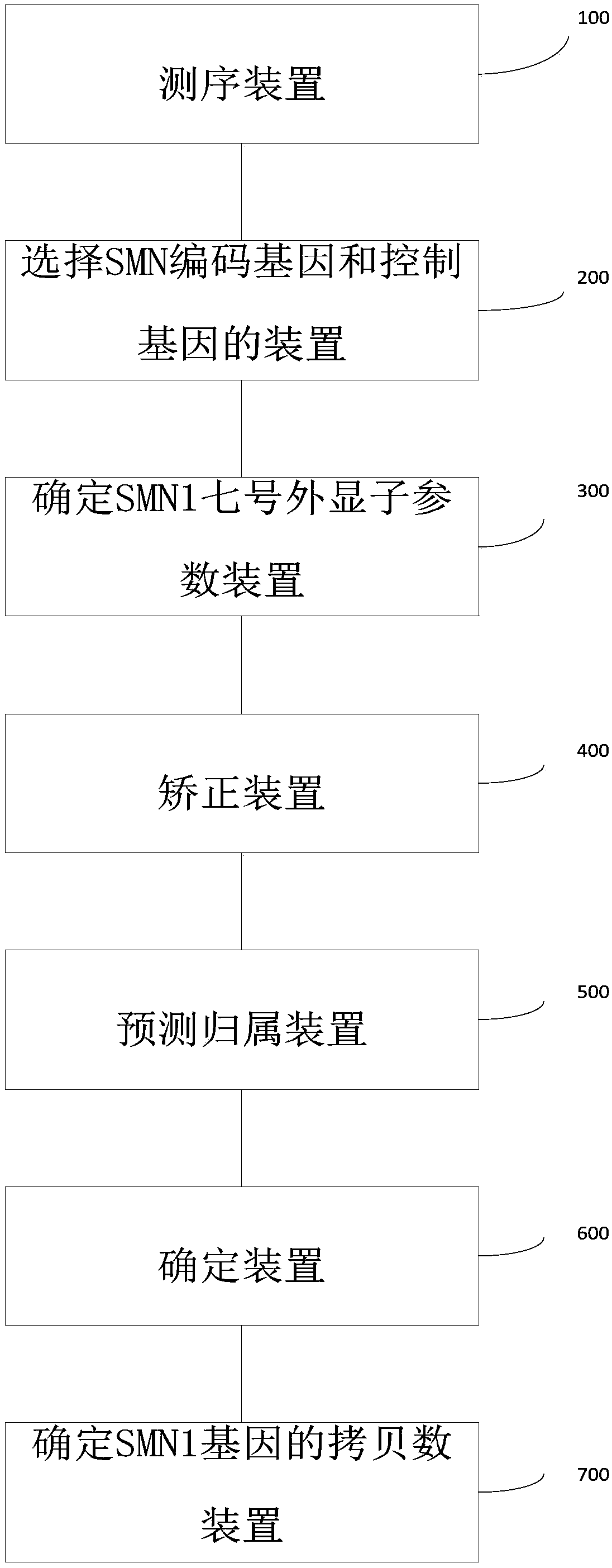
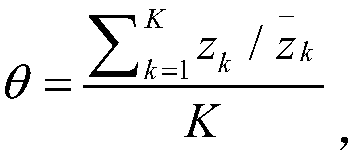Method and system for determining whether No.7 exon deletion exists in SMN1 gene of samples to be tested or not
A technology of exon deletion and exon, applied in the field of bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Embodiment 1 Original off-board data processing
[0097] Step A, apply filtering software (SOAPnuke software, version: 1.5.2) to the off-machine data to filter out low-quality sequencing reads (reads).
[0098] Step B, align the reads to the human reference genome (HG19) using alignment software (BWA software, version: 0.7.12), use Picard (a basic sequence processing software, version: 1.87) to mark repetitive sequences, and apply GATK ( A software for next-generation resequencing data analysis, version: 3.2) for re-alignment and base quality value correction.
[0099] Step C, apply the DepthofCoverage tool of GATK to obtain the reads coverage information of the capture area. It is necessary to output the locus reads number information file and the regional average coverage file (the suffix is .sample_interval_summary file).
Embodiment 2
[0100] Example 2 Estimate the copy number of exon 7 of SMN1 / 2 gene
[0101] Step A, calculate the number of reads aligned to SMN1 gene and SMN2 gene. There is only one site difference between the SMN1 gene and the SMN2 gene in the sequence of exon 7 (chr5: 70247773 on SMN1; chr5: 69372353 on SMN2, denoted as site b). Each intron has 1 site difference (chr5: 70247724 on the SMN1 gene and chr5: 69372304 on the SMN2 gene are recorded as site a, chr5: 70247921 on the SMN1 gene and chr5: 69372501 on the SMN2 gene are recorded as site c). Assuming that there are a total of N samples in the batch, calculate the ratio y of the number of SMN1 gene reads at site b to the total number of SMN reads i =B i / b i , where B i is the number of reads aligned to the b site of the SMN1 gene in the i-th sample, b i is the total number of reads compared to SMN. Also for sites a and c, calculate x i =A i / a i ,z i =C i / c i . Remember R i is the number of reads aligned to exon 7 of SM...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com