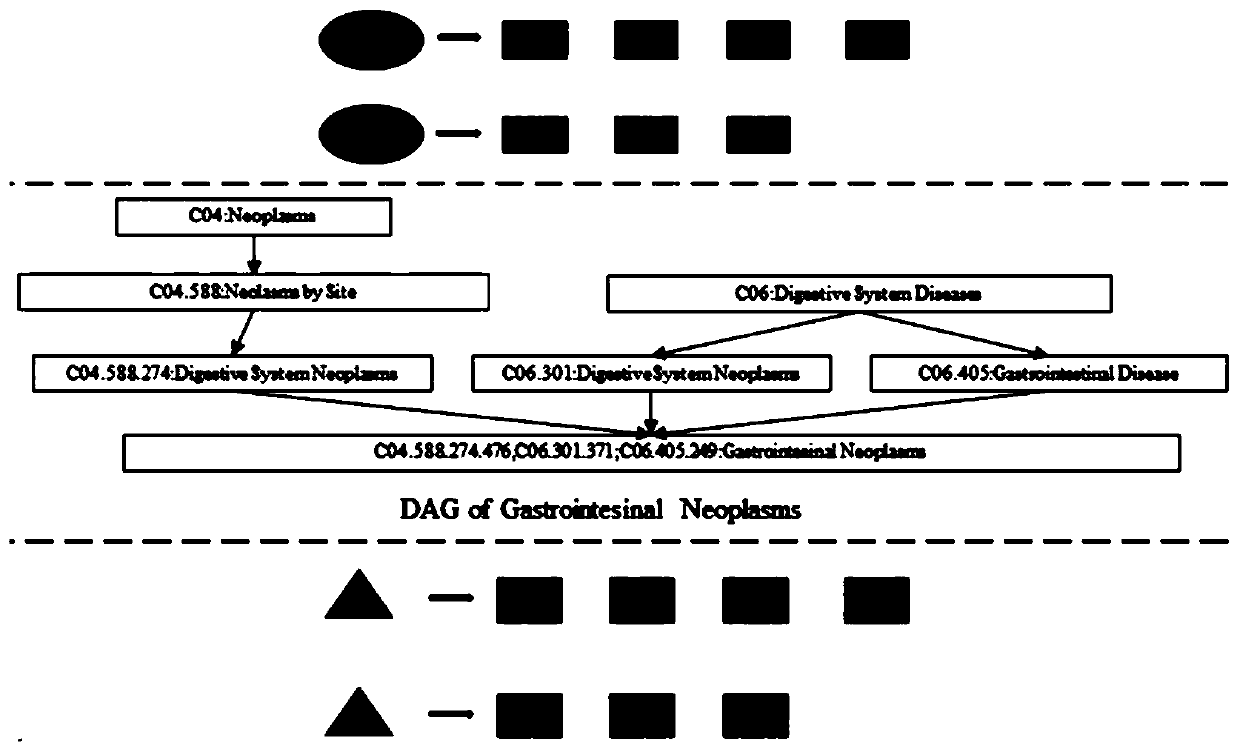
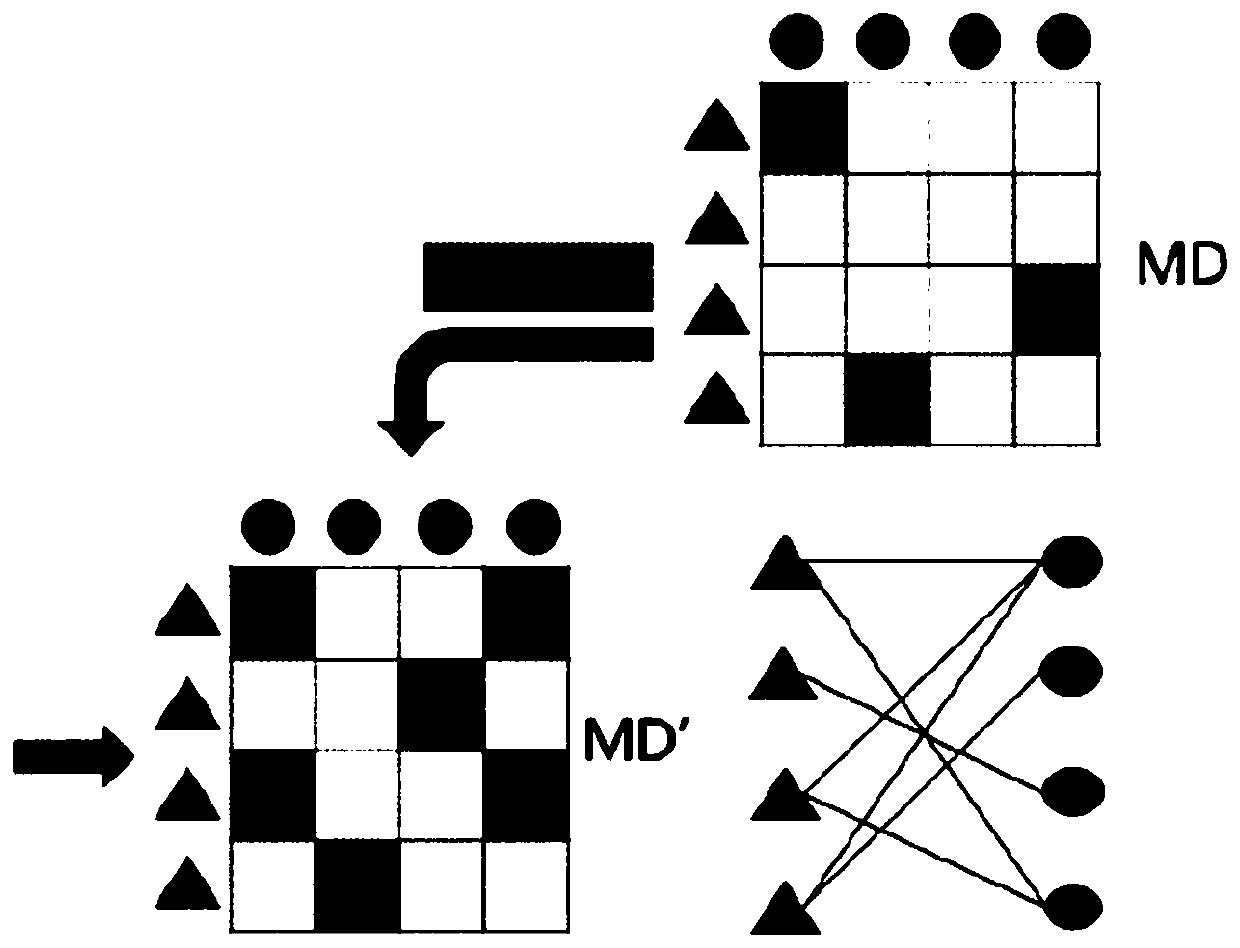
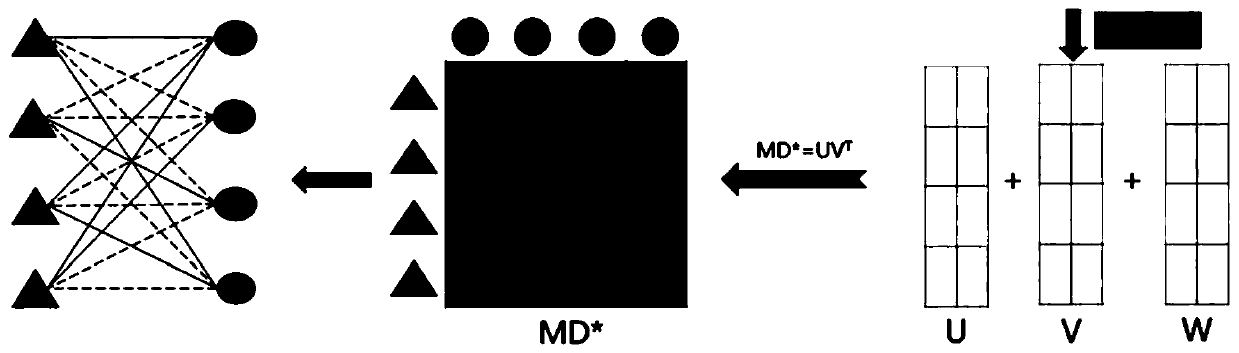
Method for predicting potential association between miRNA and disease based on constrained probability matrix decomposition
A probabilistic matrix factorization, disease technology, used in pathological reference, sequence analysis, instrumentation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0055] In order to make the purpose, technical solutions and advantages of the present invention clearer, the present invention will be described in further detail below in conjunction with experiments. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
[0056] 1. Experimental method and parameter setting
[0057] To systematically evaluate the performance of CPMDM on the collected datasets, a 5-fold cross-validation (CV) experiment was performed. In each 5-fold CV repetition, for a given disease d, the known d Related miRNAs were randomly divided into five subsets of equal size, and then one subset was used as a test set, and the remaining four subsets were used as a training set. CV experiments were performed on the training dataset to estimate the parameters. All parameter combinations are based on grid search. RMSE and AUC are used as evaluation indicators. The influe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com