Variants of cpf1 (cas12a) with altered pam specificity
A technology of S170R, cpf1, applied in DNA/RNA fragments, stable introduction of foreign DNA into chromosomes, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0372] Example 1. Variants of AsCpf1 with altered PAM specificity
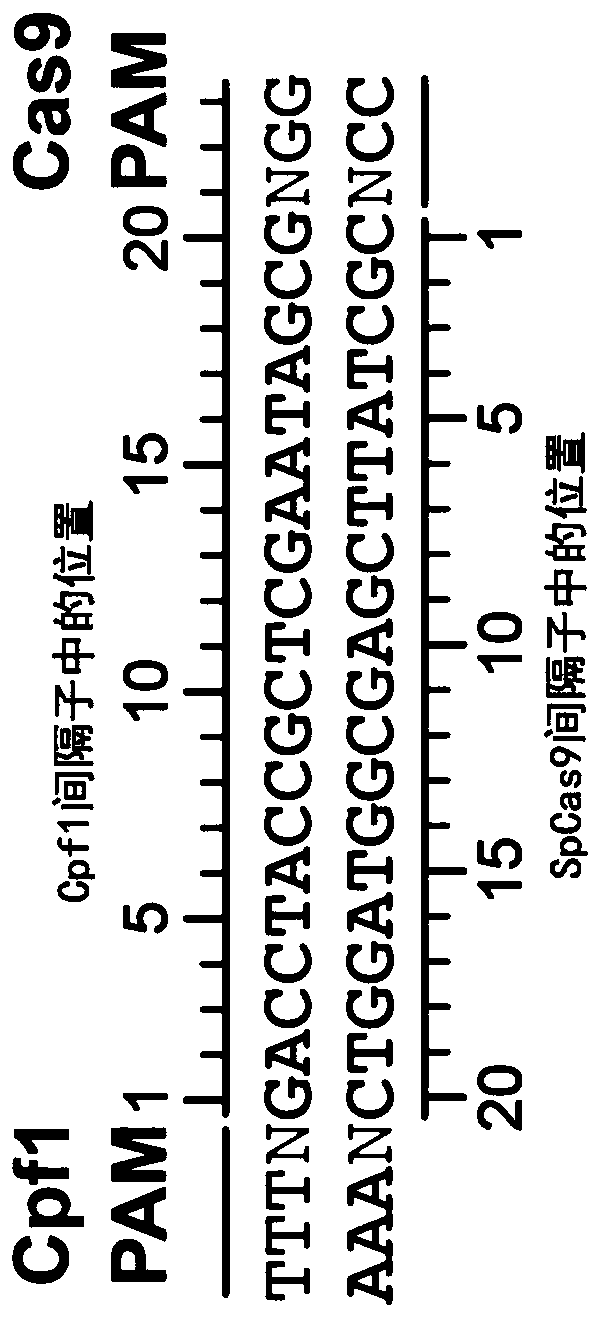
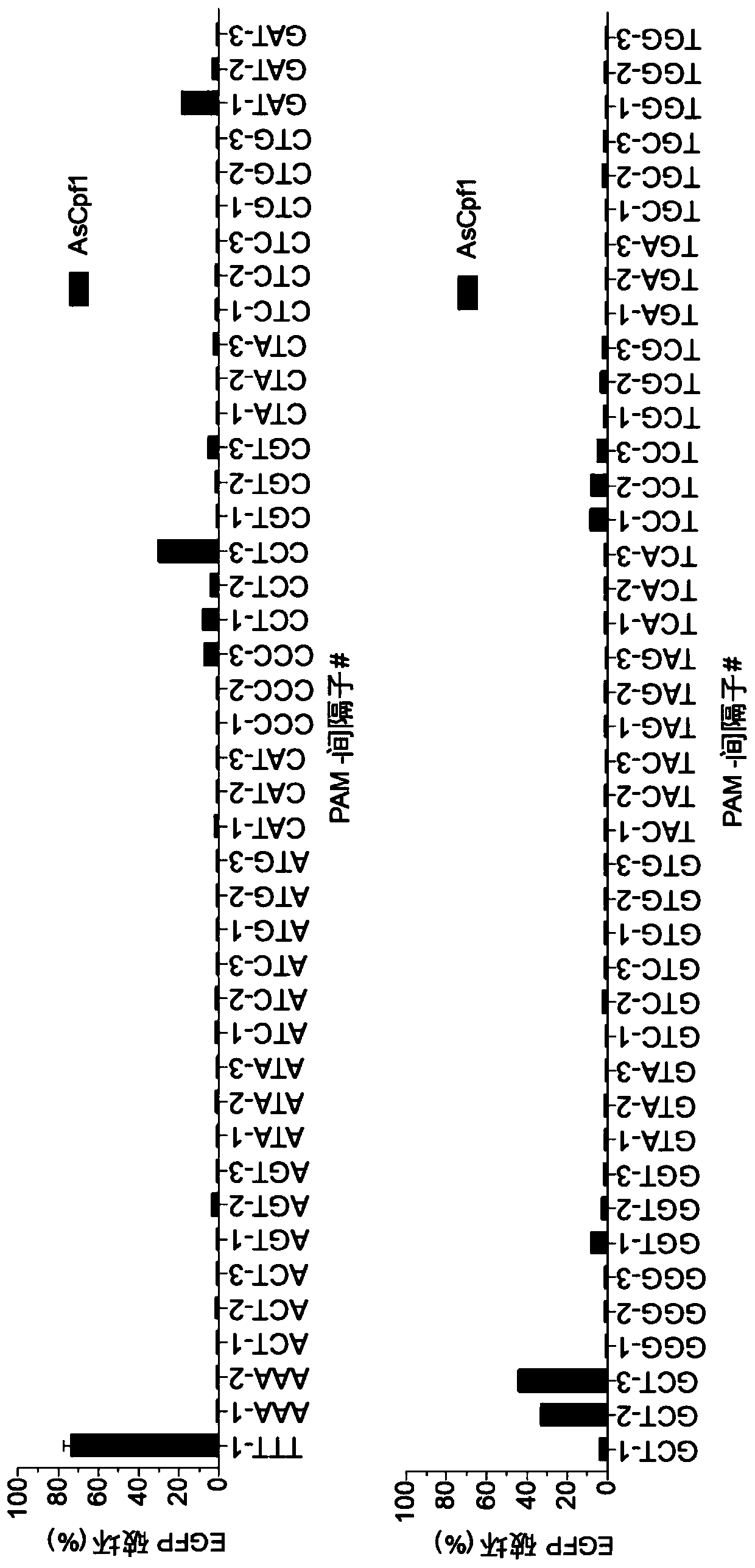
[0373] In an attempt to alter the targeting range of Cpf1 nucleases, we first examined the existing crystal structures of AsCpf1 and LbCpf1 (Dong, Nature 2016; Yamano, Cell 2016). Among other observations, these structures demonstrate that PAM specificity is mediated by a combination of electrostatic interactions and indirect base reading. We therefore hypothesized that combinations of certain amino acid substitutions at residues of DNA bases that are sterically close to PAM bases may produce variants with altered or relaxed PAM recognition preferences. To test this, we examined the region of AsCpf1 spanning residues G131-L137, S161-S181, N534-I555, Y595-T616, L628-F632 and S685-I693 near the PAM (Table 1). We focused on amino acids in the reference AsCpf1 sequence whose three-dimensional positions meet at least one of the following criteria: 1) spatially close to PAM DNA bases (on the target or non-target st...
Embodiment 1B
[0393] Example 1B. Further characterization of AsCas12a variants with altered PAM specificity and improved on-target activity
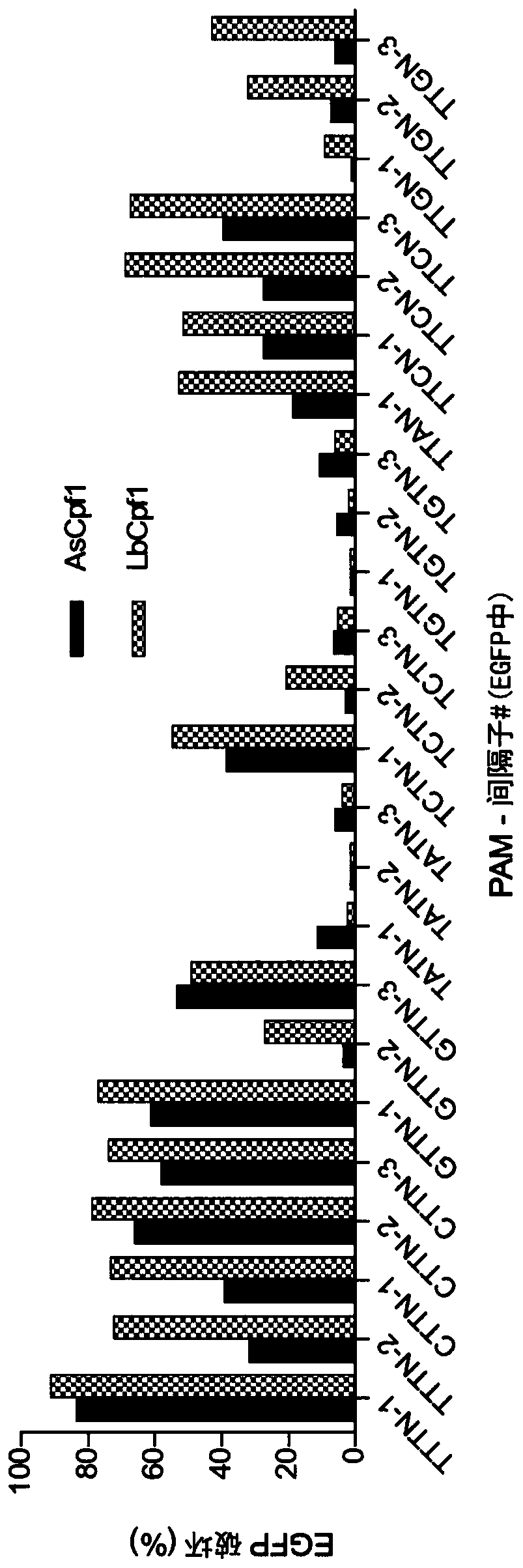
[0394] Previous characterization of Cas12a orthologs in human cells revealed that As and LbCas12a are consistently more efficient nucleases for sites with TTTV PAMs (Kim et al., Nat Biotechnol., 2016, 34:863-8) , Fn and MbCas12a may possess the relaxed PAM preference of NTTN (Zetsche et al., Cell, 2015, 163:759-71). To more thoroughly assess the activity and PAM preference of each ortholog, their genome editing activity was examined in human cells using two sets of twelve crRNAs targeting sites with TTTN or VTTNPAM ( Figure 19A ). We observed similar gene disruption among the four orthologs for the TTTN PAM site, however target-specific differences were observed. Furthermore, Fn and Mb can target VTTN PAMs more efficiently when compared with As and LbCas12a, but consistent with previous reports, their average activity towards VTTN sites is too low ...
Embodiment 4
[0408] Example 4 provides additional evidence to support the observation that the E174R substitution enhances on-target activity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com