Method for de novo assembly of genome by comprehensively applying third-generation ultra-long reads and second-generation linked reads
A genome assembly and linking technology, which is applied in the field of mixed assembly of third-generation sequencing data and second-generation sequencing data, can solve the problems of increasing sequencing and computing costs, increasing sequencing data, assembly problems and obstacles, etc., to achieve low Effects of sequencing cost and computational cost, avoidance of mismatches, and reduction of complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
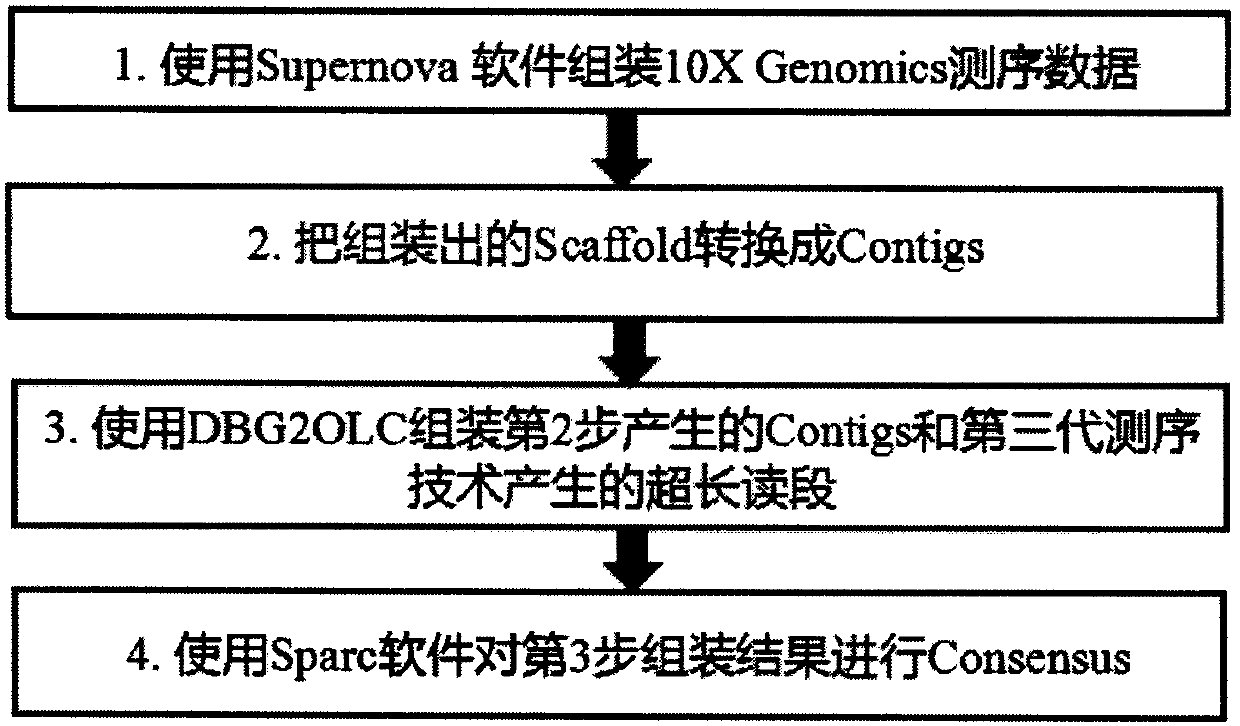
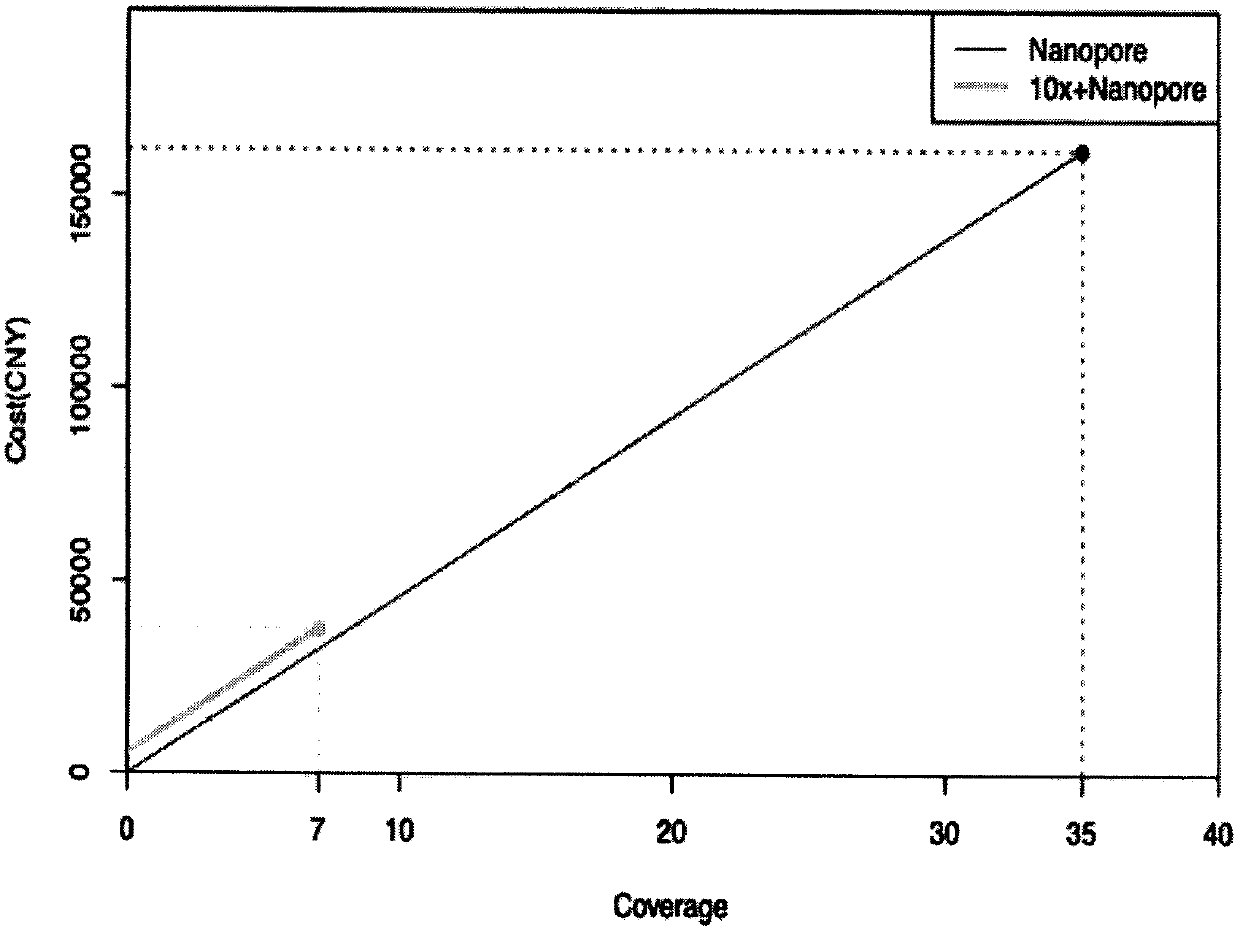
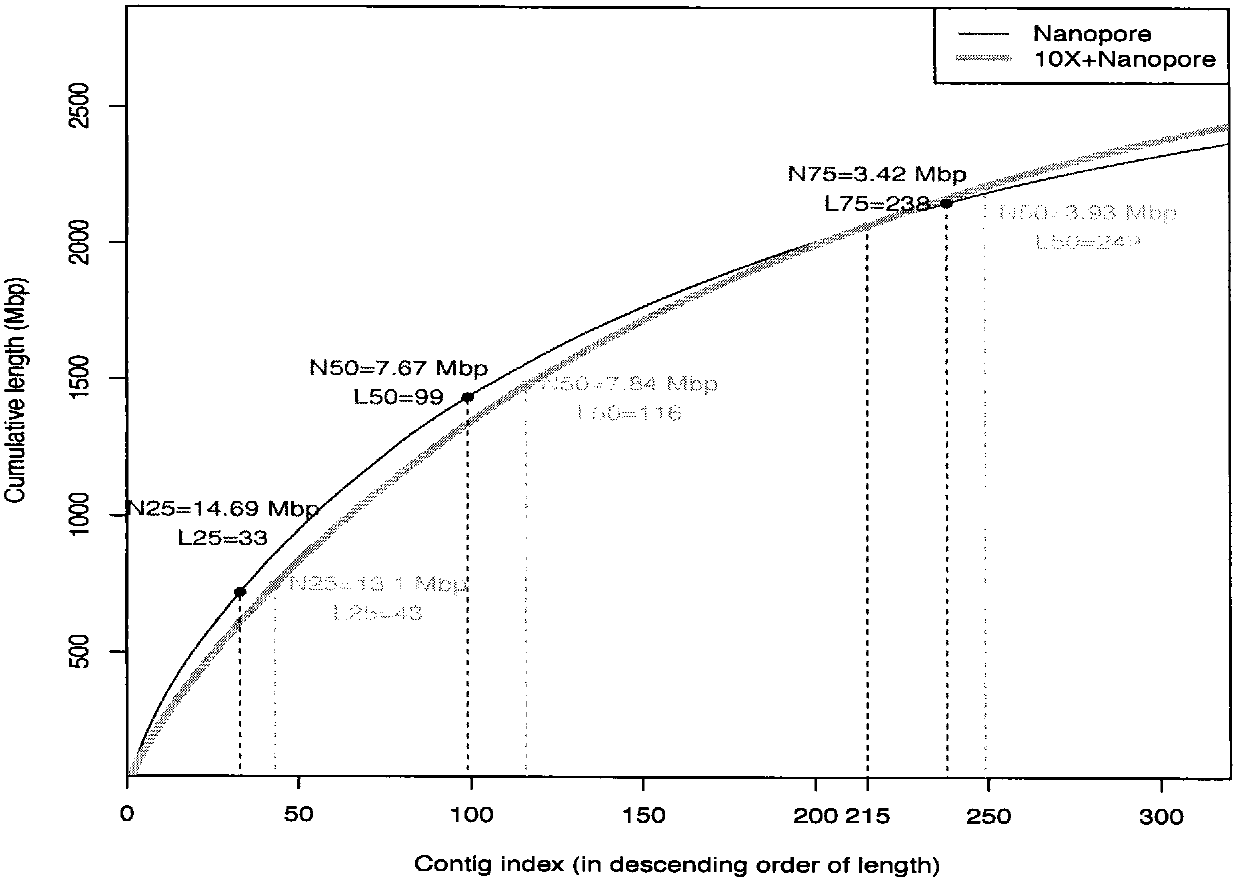
[0019] We use human genes to verify the effect of the method of the present invention. Using 56X second-generation linked read sequencing data to assemble Scaffold through Supernova, and then convert it into Contigs, and select 7X third-generation sequencing ultra-long reads. Contigs and 7X ultra-long reads were mixed assembled using DBG2OLC. In addition, we used the assembly results of the second-generation concatenated reads and the assembly results of the third-generation sequencing reads of 30X and 35X to compare the assembly effect and sequencing cost of the present invention (results are shown in Table 1).
[0020] Table 1. Comparison of 10X Genomics Linked Read Assembly Results, Nanopore Sequencing Data Assembly Results, and Hybrid Assembly Results
[0021]
[0022] **The method used in the present invention
[0023] The total length in Table 1 is the overall length of the assembled genome sequence. The total length of human is 3,000,000,000bp. The larger the value...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com