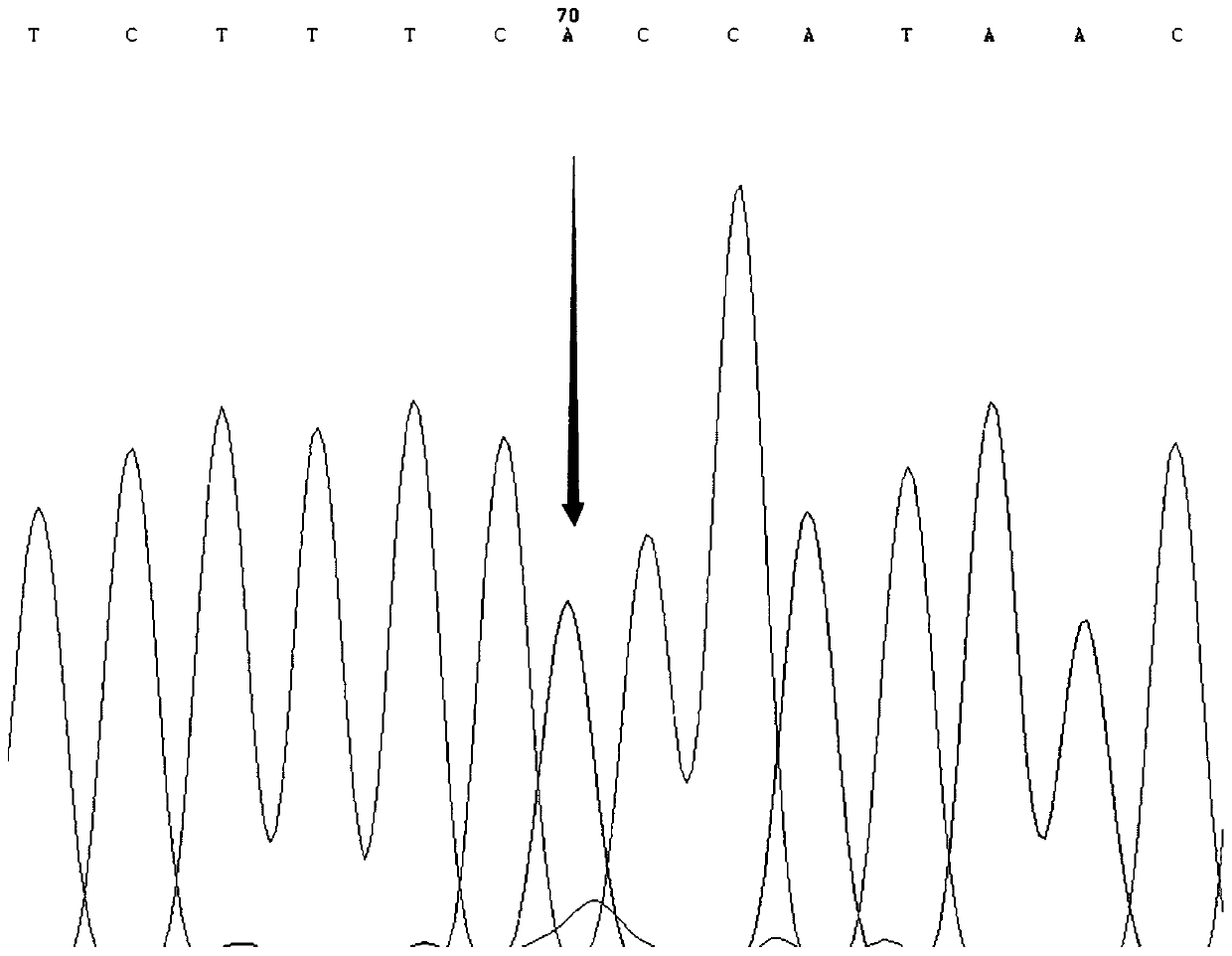
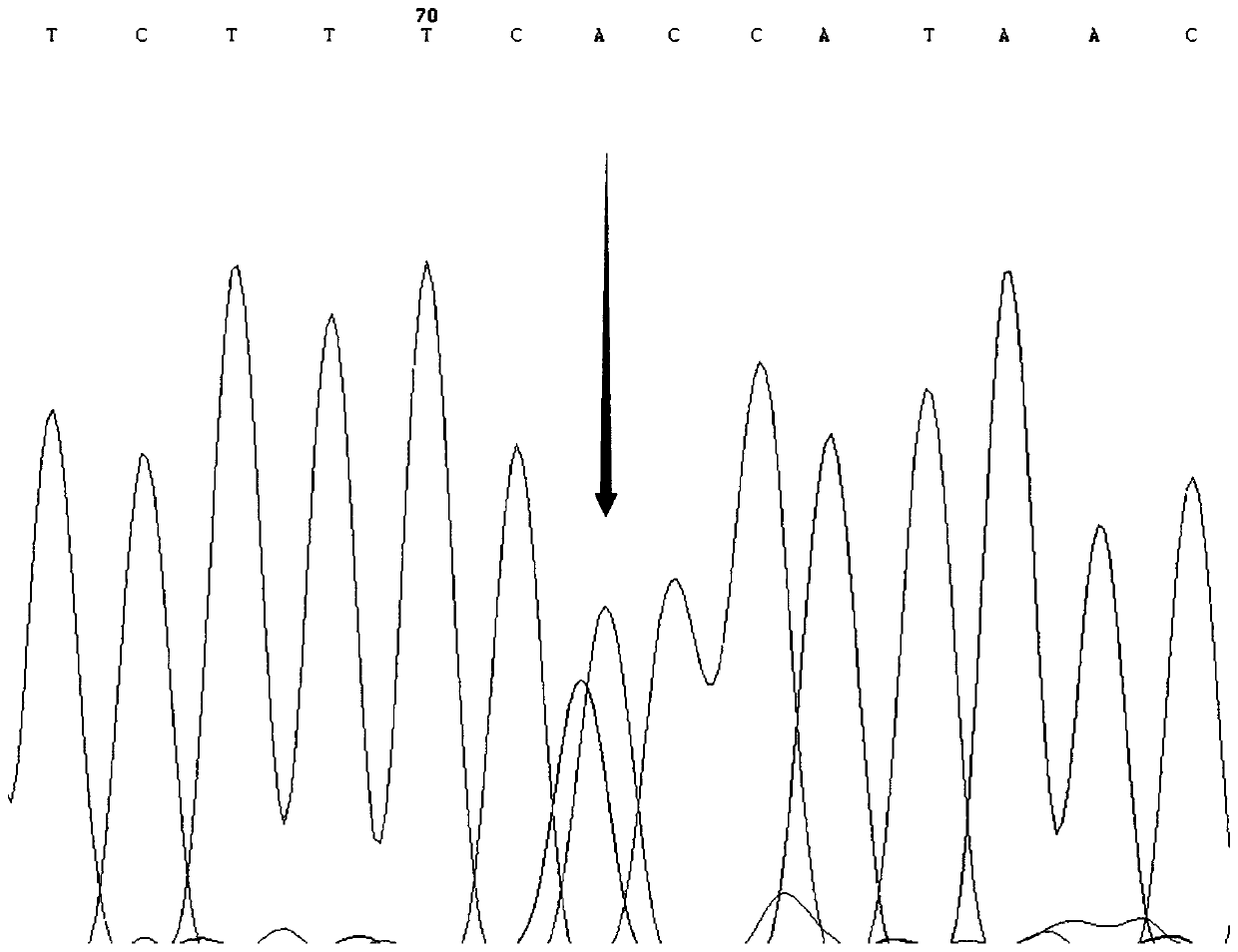
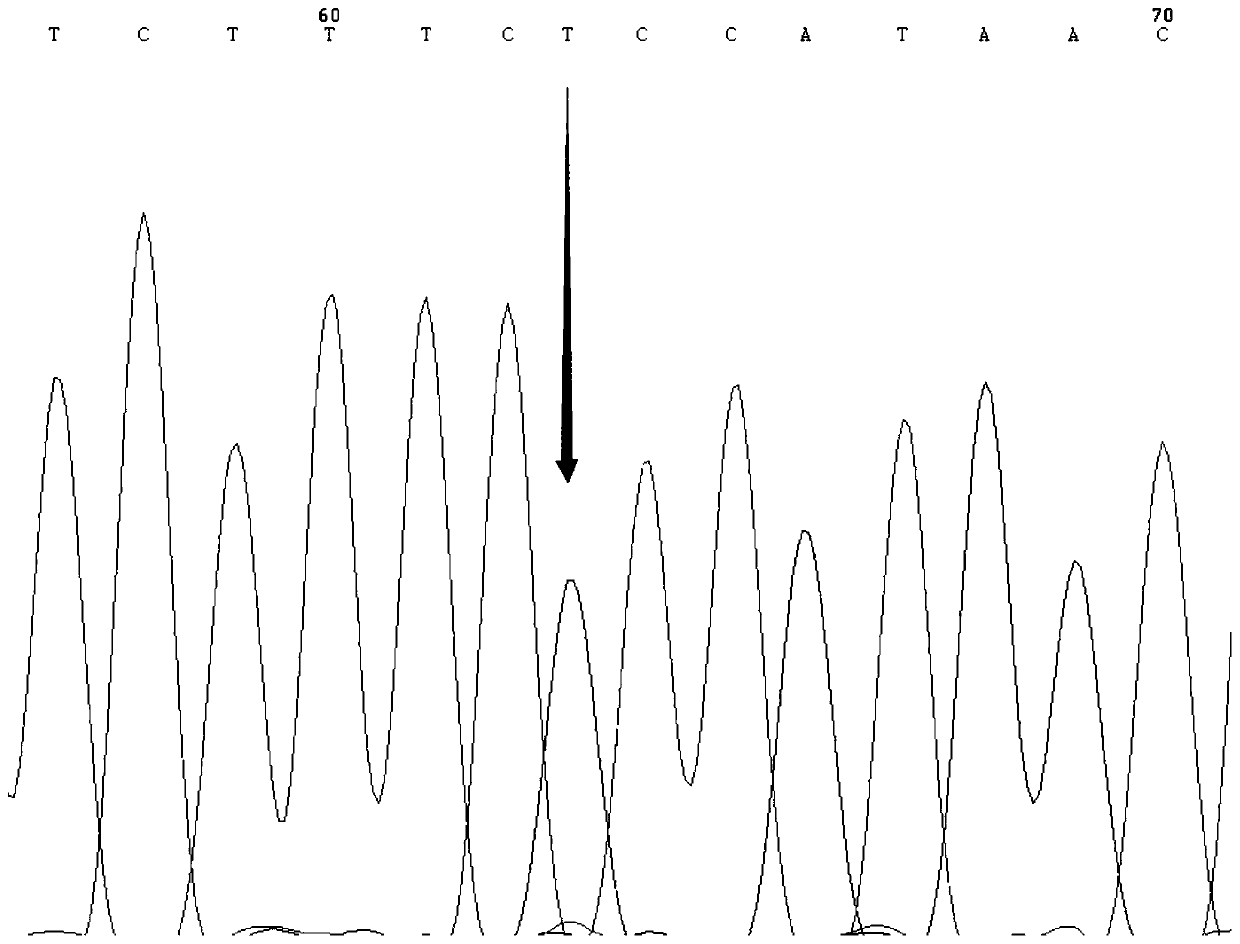
SNP molecular marker for distinguishing Takifugu fasciatus Zhongyang No. 1 from common Takifugu fasciatus and application of SNP molecular marker
A technique of puffer obscurus and molecular markers, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of CIRP combined with SNP site population identification and molecular assisted breeding applications, etc. To achieve the effect of convenient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] a) Obtaining of puffer puffer populations:
[0037]The present invention carries out the low-temperature-resistant experimental treatment on the new species of puffer puffer obscura "Zhongyang No. 1" cultivated in the Lukou Experimental Base of the Nanjing Fishery Science Research Institute. Taking the out-of-balance time as the low-temperature resistance parameter, and then performing SPSS correlation analysis on the out-of-balance time and the genotype of the SNP locus to obtain the low-temperature-resistant dominant genotype. The specific experimental process is as follows: after the film was released in late April 2019, it was cultured in the same environment (temperature, density, etc.) in the pool (580) for 191 days, and 121 tails were selected with a length of about 15 cm ± 0.5 cm. For fish with good vigor, stop feeding for 1 day before the experiment; cool down at a rate of 1°C / h from room temperature of 25°C, and cool down at a rate of 5°C / h when the temperatur...
Embodiment 2
[0064] a) Obtaining of different puffer puffer populations:
[0065] The fish used in the experiment of the present invention were taken from four groups: in September 2018, 107 pufferfish species "Zhongyang No. 1" about 5 months old were taken from Jiangsu Zhongyang Group Co., Ltd. (Hai'an); Collected from 99 common unselected Fugu obscurus populations about 4 months old in Zhenjiang Jiangzhiyuan Fishery Technology Co., Ltd. (Yangzhong) in October; in October 2018 from Jiangyin Shengang Sanxian Breeding Co., Ltd. (Jiangyin) in May There were 96 unselected puffer puffer populations around the same age; 121 fugu obscurus, a new breed of pufferfish species "Zhongyang No. 1", were bred in the Lukou Experimental Base of Nanjing Fishery Science Research Institute (Nanjing). Caudal fins were stored in 95% ethanol at -20°C for genomic DNA extraction.
[0066] b) Genomic DNA extraction of different puffer puffer populations;
[0067] The method for extracting genomic DNA from differ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com