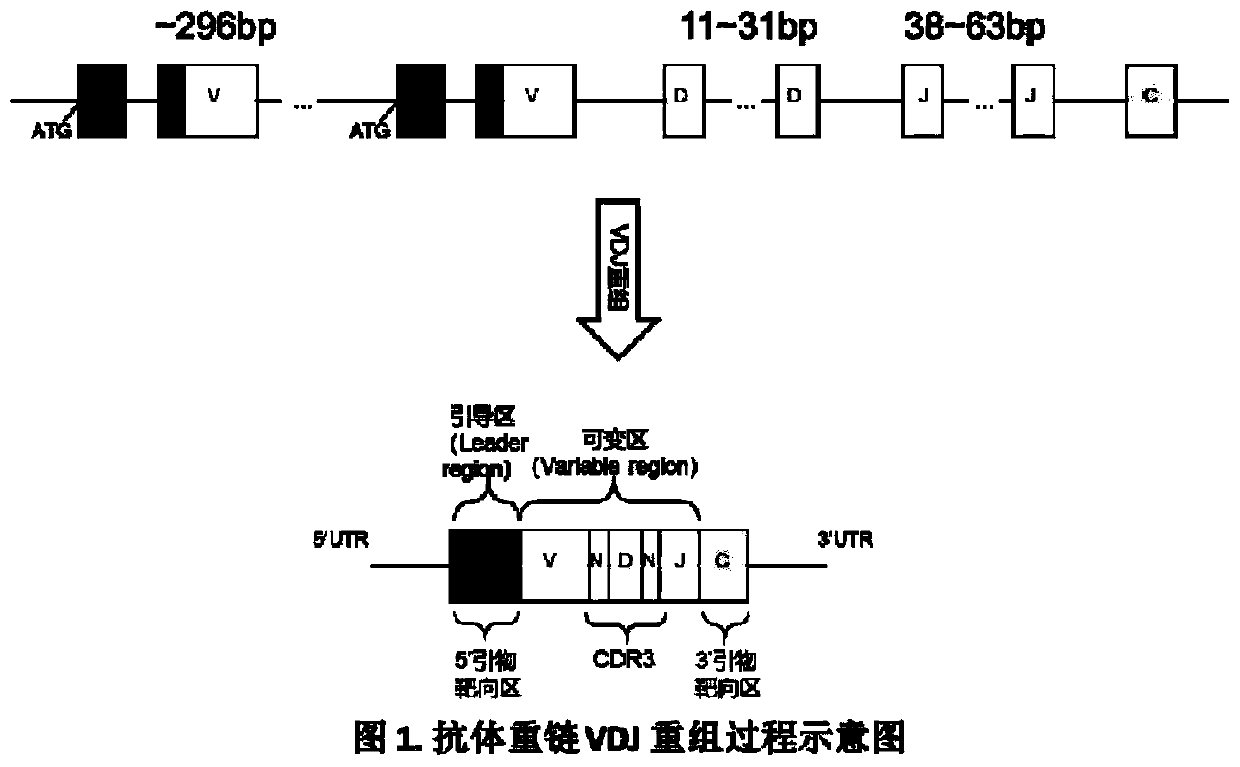
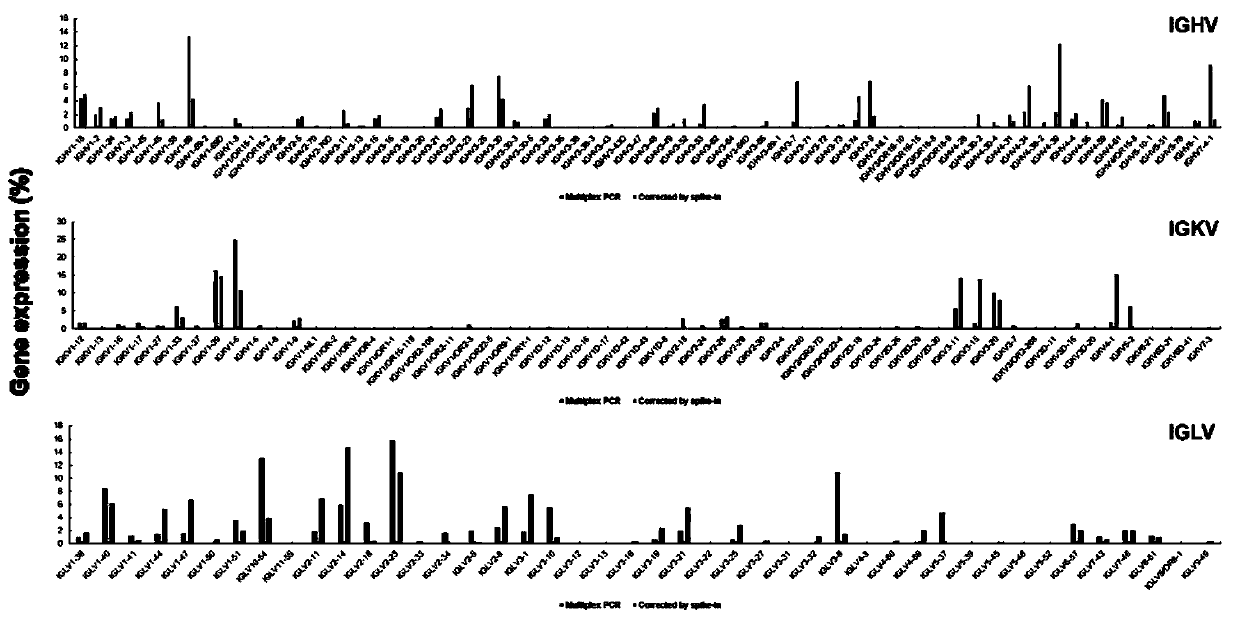
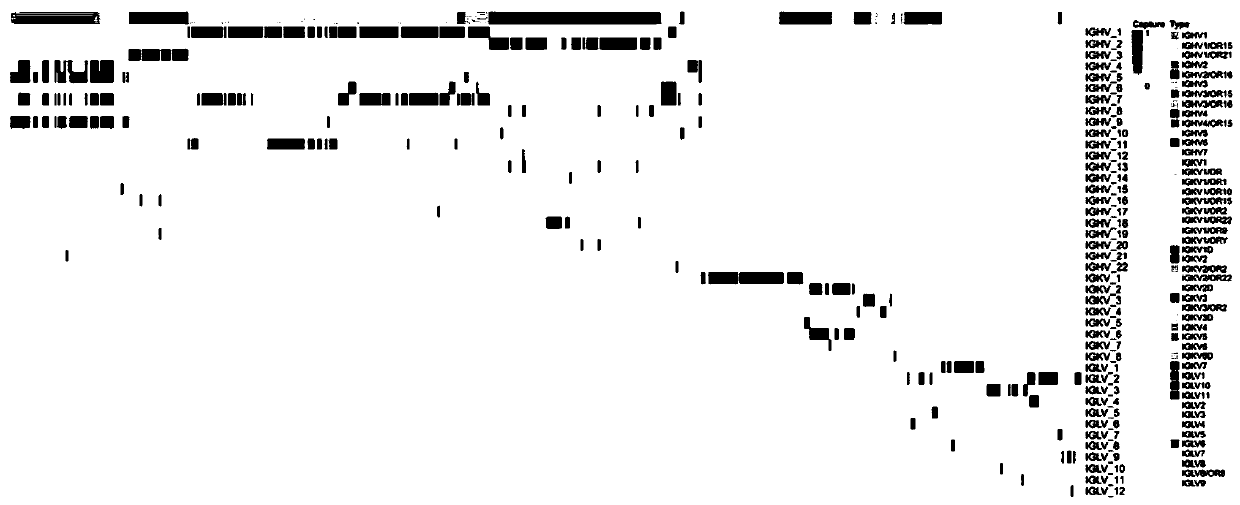Method for constructing B cell receptor (BCR) high-throughput sequencing library
A technology of sequencing library and construction method, applied in the field of biomedical services, can solve the problems of sample sequence interference, sequence error, existence, etc., and achieve the effect of correcting the preference problem
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 11
[0047] Example 1 BCR high-throughput sequencing library construction method of 1ml human peripheral blood sample
[0048] 1. Preparation of cDNA template
[0049] (1) Use EDTA anticoagulant tube to collect 1ml of human peripheral blood, use density gradient centrifugation to separate mononuclear cells in the blood, add 600ul cell lysate for cell lysis, use RNA extraction kit for RNA extraction; (2) detect RNA (3) Configure the mixed system according to the following table 5, and carry out the reaction on the PCR instrument. The reaction program is: 70 ° C, 1 min. After the reaction program is completed, immediately put the product on ice for 3 min. table 5
[0050]
[0051] (4) Configure the mixing system in Table 6 below, mix it with the product of step (3) and put it on the PCR instrument for reaction. The reaction program is: 42°C, 90min, 75°C, 15min, 4°C, ∞. The product after the reaction is the synthesized cDNA template C, which will be used in the next experiment. ...
Embodiment 2
[0064] Example 2: BCR high-throughput sequencing library construction of 1 mg human breast cancer tissue sample
[0065] 1. Preparation of cDNA template: (1) Take out human breast cancer tissue sample from liquid nitrogen or -80°C refrigerator, weigh 1 mg, grind the tissue into powder by liquid nitrogen grinding method, add 600ul cell lysate for cell lysis , use the RNA extraction kit to extract RNA; (2) Detect the integrity of RNA and measure the concentration of RNA; (3) Configure the mixed system according to the following table 10, and perform the reaction on the PCR machine. The reaction program is: 70°C , 1min, immediately put the product on ice for 3min after the completion of the reaction procedure. Table 10
[0066]
[0067] (4) Configure the mixing system in Table 11 below, mix it with the product of step (3) and put it on the PCR instrument for reaction. The reaction program is: 42°C, 90min, 75°C, 15min, 4°C, ∞. The product after the reaction is the synthesized...
Embodiment 3
[0083] Through the analysis of the respective primer sets of IgG, IgL, and IgK, we can see that the three sets of primers can amplify all alleles of IgG, IgL, and IgK currently known ( figure 2 ):
[0084] Such as figure 2 : This figure is a heat map of the corresponding relationship between primers and amplified genes. The abscissa represents each allele, and the ordinate represents each primer sequence. If the primer can amplify the allele, the color is red, otherwise it is white. It can be seen from this figure that the sequence of this set of primers has strong specificity, that is, the heavy chain primers only amplify the alleles corresponding to the heavy chain, and the light chain primers only amplify the alleles corresponding to the light chain. Secondly, this set of primers can amplify all alleles, which can effectively measure the diversity of BCR.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com