Method for preparing nucleic acid for sequencing and application of nucleic acid
A sequencing and nucleic acid technology, applied in genetic testing and biological fields, can solve problems affecting the structure of the sequencing library, slow down the running speed, and the construction method of the sequencing library needs to be optimized, so as to achieve the effect of good library structure and large output of sequencing data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] 1. Utilize the method of the embodiment of the present invention to construct a sequencing library, the method is as follows:
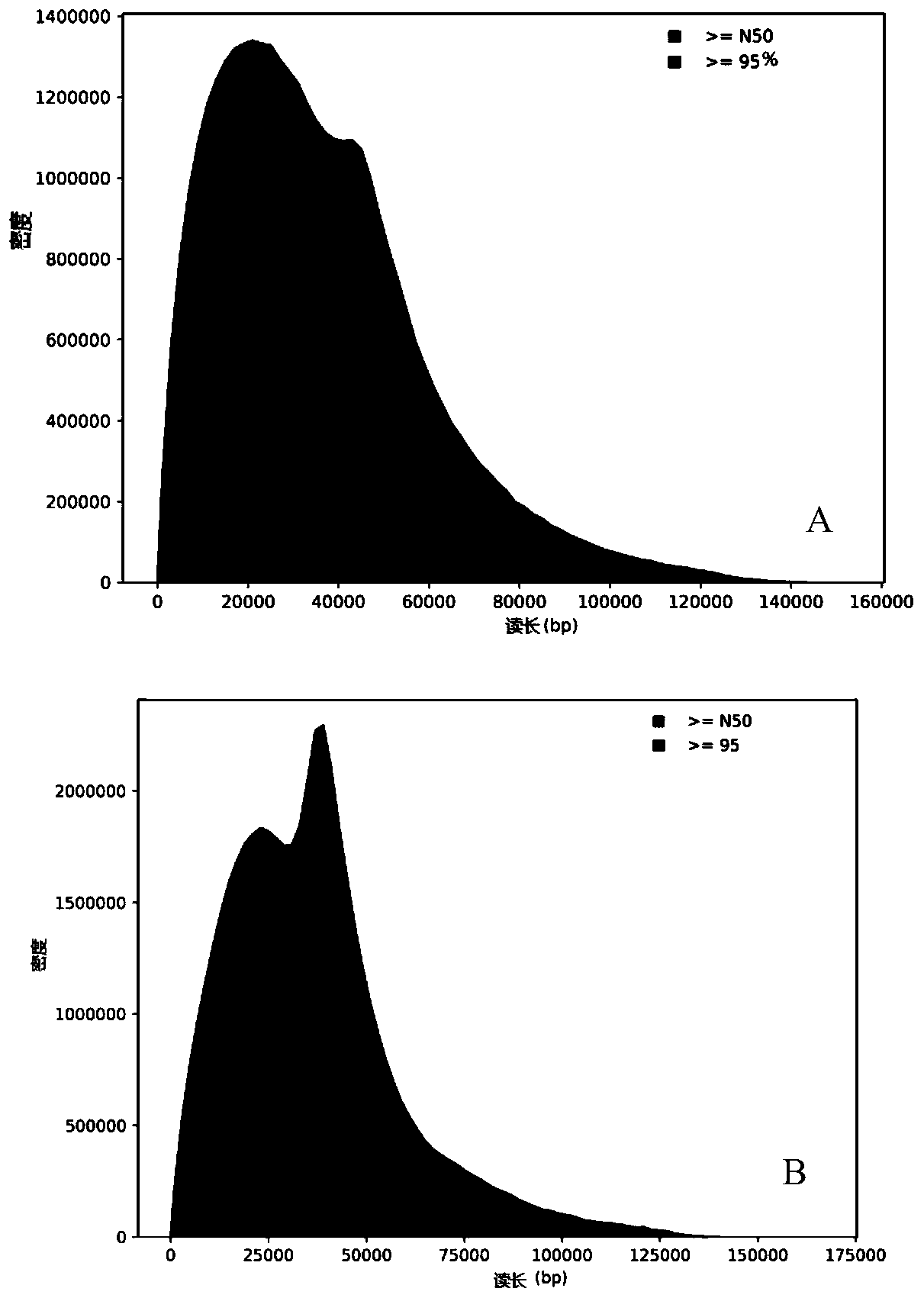
[0069] (1) Take 8 μg of extracted mouse lung genomic DNA, cut it into large fragments of 30-40K, perform end repair and add A treatment, and the reaction reagents include T4 DNAP, T4 PNK, HemoklenTaq and universal buffer.
[0070] (2) The genome end repair product was ligated with the lox66 sequence coupled with cohesive ends, and the reaction reagents included T4 DNA ligase and Transformer buffer.
[0071] (3) The ligation product was purified using Ampure purification magnetic beads to obtain the purified product.
[0072] (4) UDG cleavage and circularization of the ligation product, the reagents include but not limited to T4 DNA ligase, UDG enzyme and matching buffer.
[0073] (5) Digestion, the reagents include T4 exonuclease 7 and matching buffer.
[0074] (6) Site-specific recombination of the digestion product with the hetero-adapter, ...
Embodiment 2
[0097] 1. Utilize the method of the embodiment of the present invention to construct a sequencing library, the method is as follows:
[0098] (1) Take 8ug of extracted mouse lung genomic DNA, cut it into large fragments of 10-15K, and perform end repair and A treatment. The reaction reagents include T4 DNAP, T4 PNK, HemoklenTaq and universal buffer.
[0099] (2) The genome end repair product was ligated with the lox66 sequence coupled with cohesive ends, and the reaction reagents included T4 DNA ligase and Transformer buffer.
[0100] (3) The ligation product was purified using Ampure purification magnetic beads to obtain the purified product.
[0101] (4) UDG cleavage and circularization of the ligation product, the reagents include but not limited to T4 DNA ligase, UDG enzyme and matching buffer.
[0102] (5) Digestion, the reaction reagents include T4 exonuclease 7 and supporting buffer.
[0103] (6) Site-specific recombination of the digestion product with the hetero-ada...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com