Optimum design method of structure-based CRISPR protein
An optimized design and protein technology, applied in the field of protein engineering, can solve problems such as time-consuming, labor-intensive, blindness, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0048] The experimental methods used in the following examples are conventional methods unless otherwise specified.
[0049] The materials and reagents used in the following examples are obtained from commercial sources unless otherwise specified.
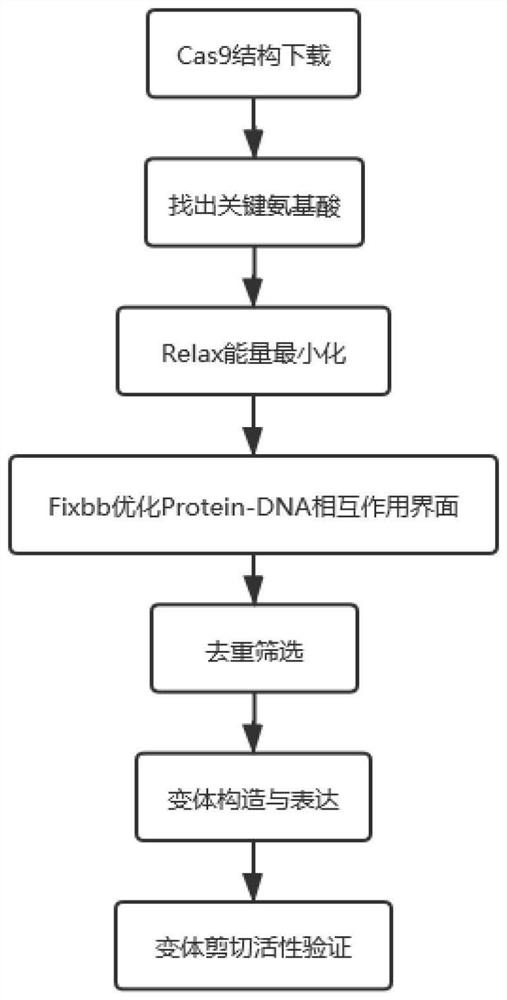
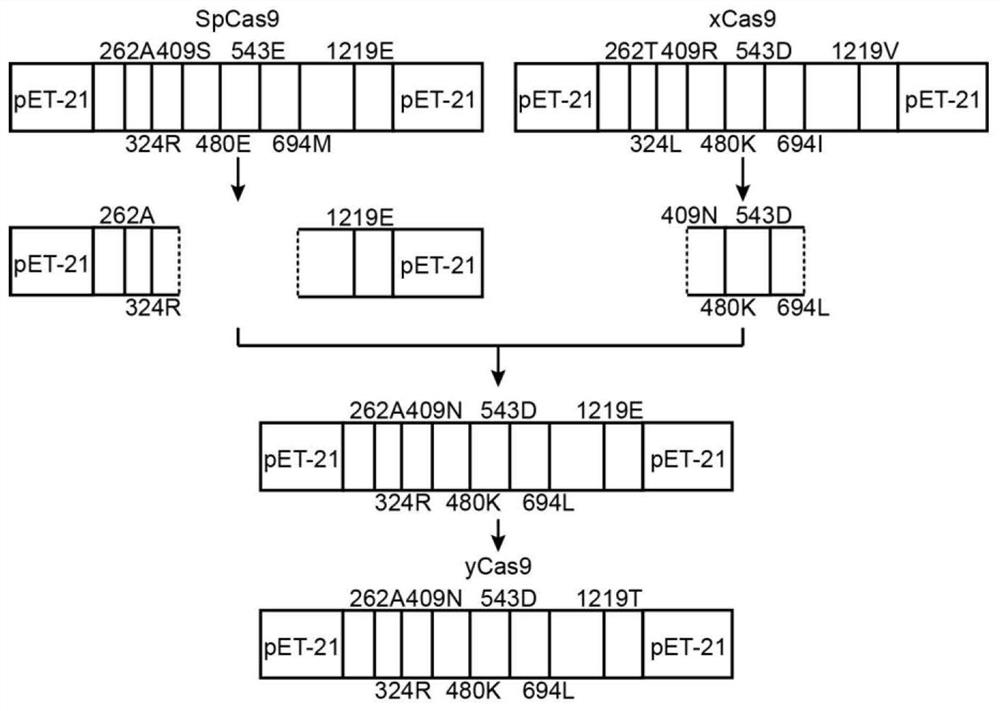
[0050] 1. SpCas9 combinatorial mutation design
[0051] Step 1: Identify the amino acid positions that have an important impact on the properties of Cas9.
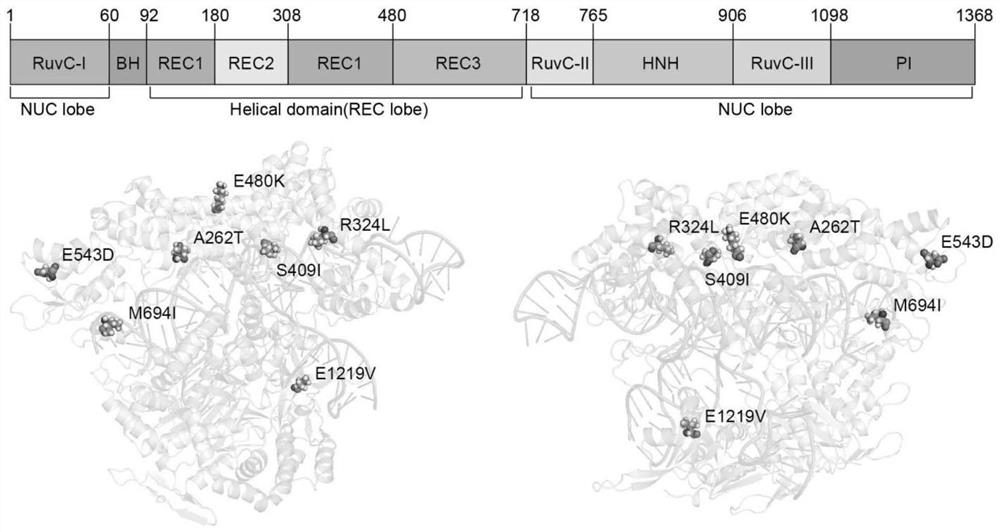
[0052] The seven directed evolution sites (A262T, R324L, S409I, E480K, E543D, M694I, and E1219V) of xCas9 mutations are distributed on both sides of the sgRNA-DNA heteroduplex (attached figure 2 ). These seven amino acids work together to expand the PAM recognition range of xCas9 and reduce its off-target effects. Therefore, these 7 sites were selected as the amino acid sites for Rosetta optimization design.
[0053] Step 2: Minimize Relax energy.
[0054] The SpCas9 structure (PDB ID: 4UN3) was iterated for 25 rounds with the Relax module, and the structure with the lowe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com