PyrG screening marker recycling method and application
A screening marker, recombinant bacteria technology, applied in the biological field, can solve the problems of repetitive sequence residues, limited genetic screening markers, etc., and achieves the effect of simple operation and wide application range.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
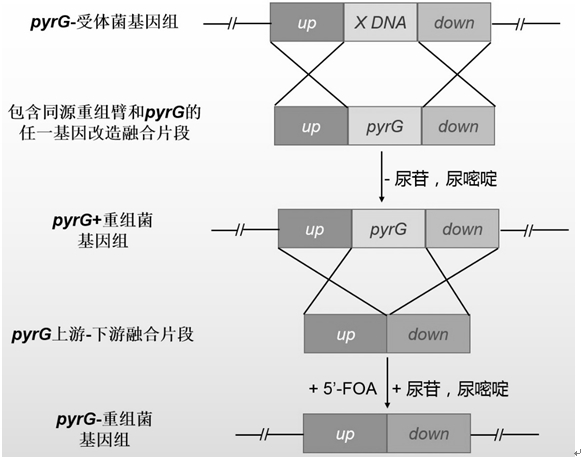
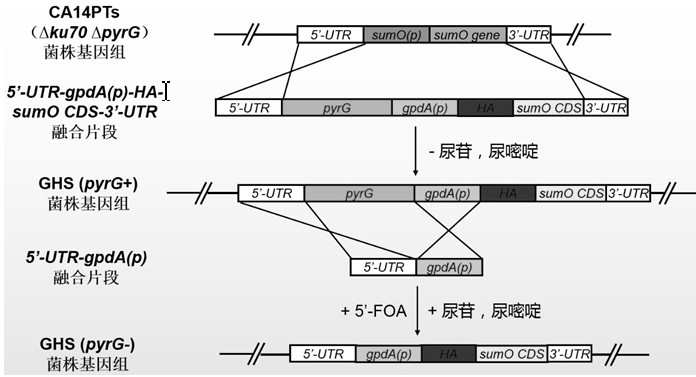
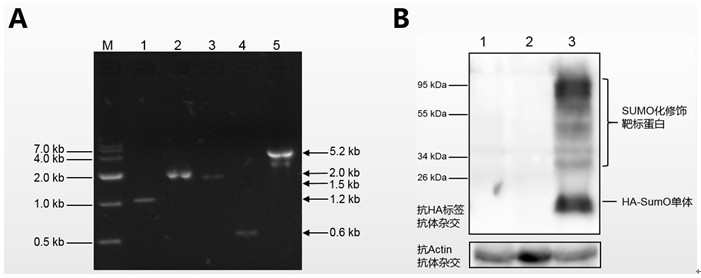
[0030] The inventor established a new orotidine-5'-phosphate decarboxylase gene after careful research pyrG A method for the recycling of genetic selection markers. This method requires only a pyrG The recycling of genetic screening markers can realize multiple genetic transformation operations, avoiding the impact of differences in different genetic screening markers on the study of fungal genetic background and traits, and using this method to eliminate pyrG Genetic screening markers do not need to rely on direct repeats, knockout pyrG There will be no repetitive sequences remaining on the genome after the screening markers, and it will not affect the genetic background and traits of the recombinant bacteria, which can solve the problem of the limitation of genetic screening markers in the process of fungal genetic transformation and the impact of different genetic screening markers on the study of fungal genetic background and traits and traditional URA3 / pyrG The repeat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com