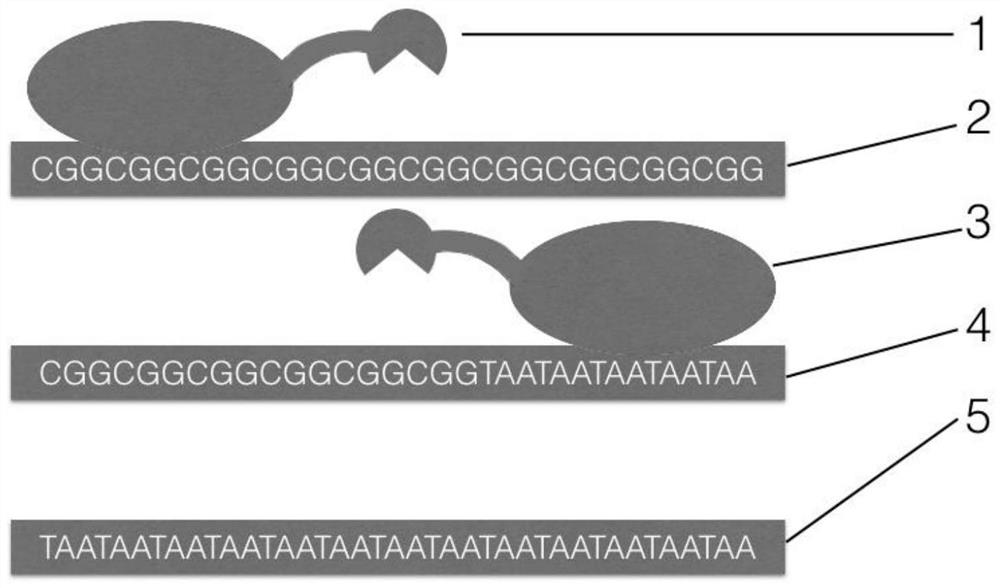
Amplification method for analyzing CGG repeat number of upstream untranslated region of fragile X mental retardation 1 (FMR1) gene
A technique of untranslated regions and repeat numbers, applied in the field of medical laboratory science, which can solve problems such as accuracy impact
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0040] Example: Using the amplification method for analyzing the number of CGG repeats in the upstream untranslated region of the FMR1 gene to detect the number of CGG repeats in the upstream region of the upstream untranslated region of the human FMR1 gene.
[0041] (1) Prepare experimental components and experimental equipment
[0042] (1) Components required for making experiments:
[0043] Preparation of recombinant TALE-deaminase: clone the gene sequence of TALE-deaminase into E. coli expression vector PET28a;
[0044] Wherein, CGG-TALE-downstream deaminase expression sequence (including start codon and stop codon):
[0045] ATGCATGGCCTGACTCCGGACCAAGTGGTGGCTATCGCCAGCCACGATGGCGGCAAGCAAGCGCTCGAAACGGTGCAGCGGCTGTTGCCGGTGCTGTGCCAGGACCATGGCCTGACCCCGGACCAAGTGGTGGCTATCGCCAGCAACAAGGGCGGCAAGCAAGCGCTCGAAACGGTGCAGCGGCTGTTGCCGGTGCTGTGCCAGGACCATGGCCTGACCCCGGACCAAGTGGTGGCTATCGCCAGCAACAAGGGCGGCAAGCAAGCGCTCGAAACGGTGCAGCGGCTGTTGCCGGTGCTGTGCCAGGACCATGGCCTGACTCCGGACCAAGTGGTGGCTATCGCCAGCCACG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com