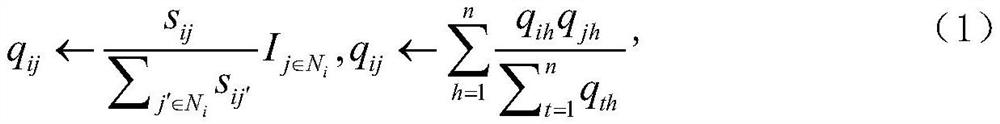Fusion network drug target relationship prediction method based on network enhancement and graph regularization
A technology that integrates networks and prediction methods, applied in the field of systems biology, can solve the problems of low prediction accuracy, incomplete extraction of molecular information, and large noise in biological data, and achieve the effect of suppressing noise.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
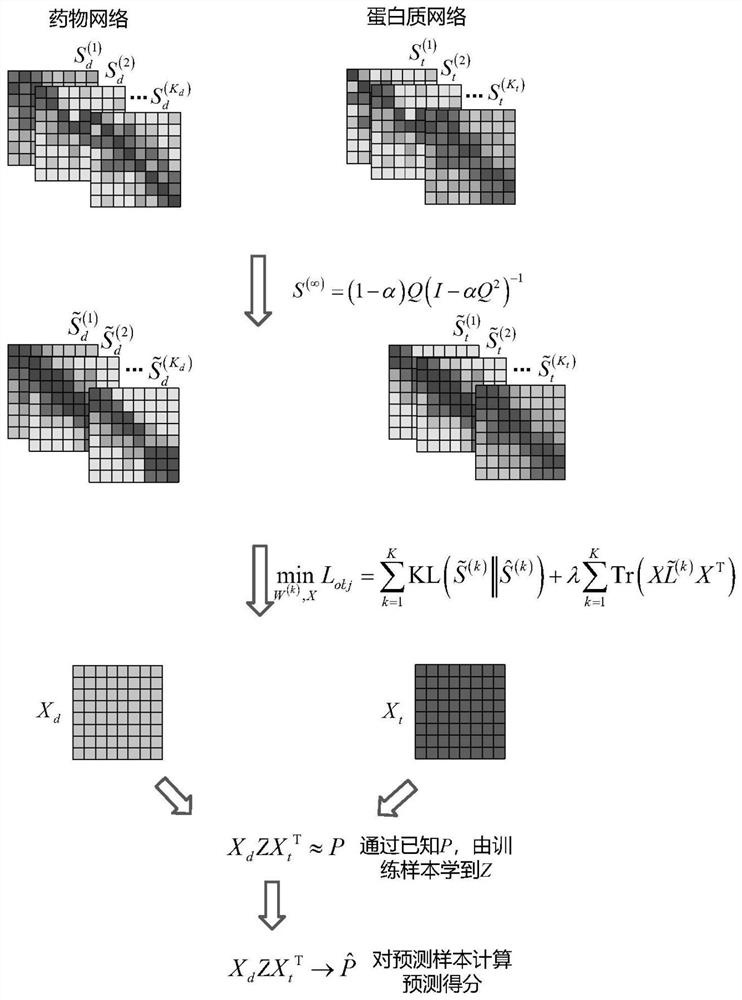
[0056] Such as Figure 1-Figure 2 As shown, a fusion network drug target relationship prediction method based on network enhancement and graph regularization includes the following steps:
[0057] S1: Using the undirected graph model to model the drug similarity network and protein similarity network respectively;
[0058] More specifically, the drug similarity network includes: drug chemical structure similarity, drug-disease association similarity, drug-side effect similarity, drug interaction similarity; the protein similarity network includes: sequence structure similarity, protein- Disease association similarity, protein interaction similarity.
[0059] S2: Use the network enhancement method based on the third-order neighborhood random walk to enhance the modeled drug similarity network and protein similarity network;
[0060] The specific process includes:
[0061] S201: Both the drug similarity network and the protein similarity network are represented by G, N repres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com