A circular RNA molecule for detecting radiosensitivity of rectal cancer and its application
A sensitive and colorectal cancer technology, applied in the field of clinical medicine, to achieve high specificity, optimized treatment strategy, and accurate detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1 Screening of differentially expressed circRNAs
[0047] Modeling 12 rectal cancer patients, 5 of them achieved pathological complete remission after preoperative radiotherapy (radiotherapy sensitive group, code G), and 7 cases were postoperative pathologically graded 3-4 (Mandard grading standard, radiotherapy resistant group, code U). Clinical samples of colorectal cancer tissues from each patient were collected, and total RNA was extracted from all clinical sample tissues by the trizol method and qualified for quality control before library construction and sequencing. Specific steps are as follows:
[0048] (1) Extract total RNA:
[0049] Take the tissue out of the liquid nitrogen, transfer it to a 1.5mL RNase-free EP tube, add 1ml Trizol lysate immediately, use a tissue homogenizer to grind and lyse the tissue cells, and wait until the liquid is cloudy and there is no obvious tissue block at 12,000rpm Centrifuge for 5 minutes, and transfer the supernata...
Embodiment 2
[0054] Example 2 qRT-PCR sequencing verification
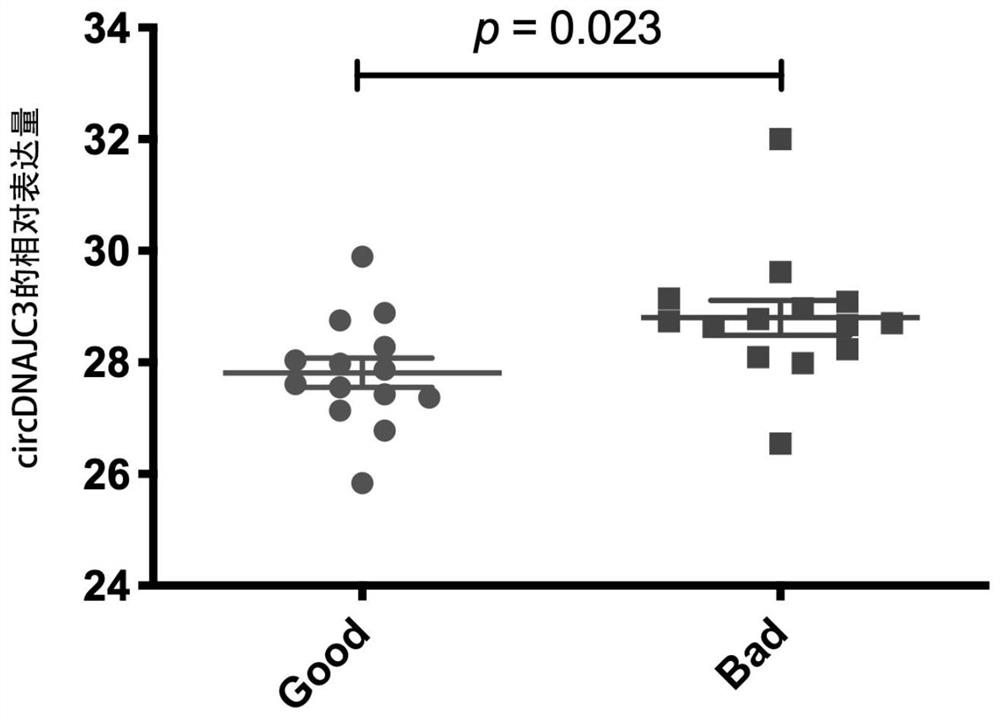
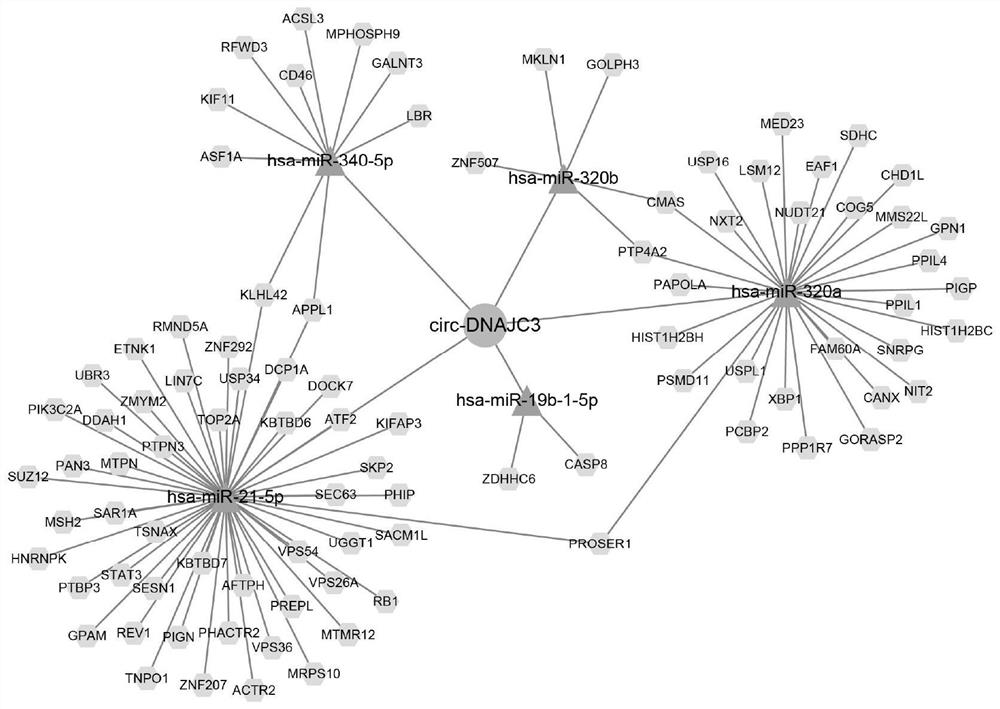
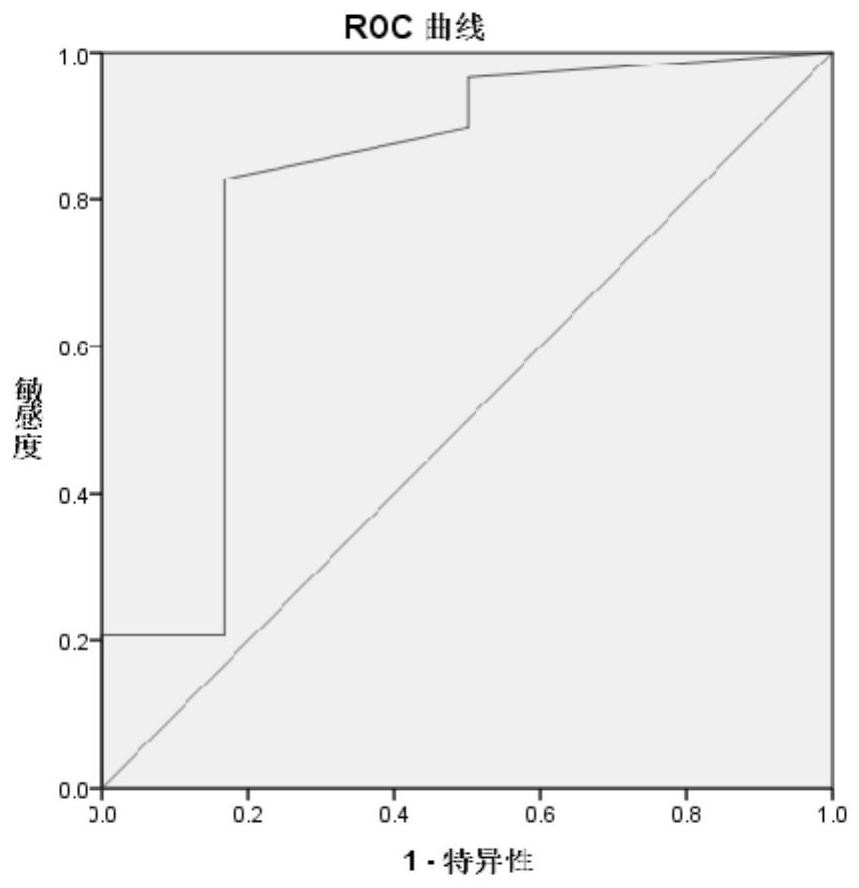
[0055] Based on the 119 circRNAs with significantly different expression levels obtained in Example 1, circDNAJC3 among them was selected for verification in this example. Specific steps are as follows:
[0056] (1) Extract tissue RNA
[0057] According to the method of Example 1, the clinical tissues of 28 patients (including 14 cases in the radiotherapy-sensitive group and 14 cases in the radiotherapy-resistant group) who had undergone preoperative radiotherapy were collected, and RNA was extracted respectively.
[0058] (2) cDNA synthesis
[0059] 1. RNA pre-denaturation:
[0060] Add the following reaction liquids to the microcentrifuge tube, as shown in Table 1:
[0061] Table 1
[0062] Reagent Dosage RNA 2.0μg Random / Oligo primer 1μL wxya 2 o
Make up to 16μL
[0063] Incubate at 65°C for 5 minutes, and immediately place the mixture on ice for 2 minutes;
[0064] 2. RNA rev...
Embodiment 3
[0084] Example 3 Transcriptome sequencing verification
[0085] This embodiment is further extended to 35 independent specimens (including 6 cases in the radiotherapy-sensitive group and 29 cases in the radiotherapy-resistant group) for transcriptome sequencing, and the specific steps are as follows:
[0086] One: miRNA sequencing experiment process
[0087] (1) Nucleic acid extraction: collect and extract RNA according to the method of Example 1
[0088] (2) Library construction
[0089] 1. Reagents: Multiplex Small RNA Library Prep Kit
[0090] 2. Instrument: 2720Thermal Cycler (life Technologies), Agilent Technologies 2200 TapeStation (life Technologies)
[0091] 3. miRNA library construction steps:
[0092] 3.1 3' end connector connection:
[0093] Take total RNA 100ng-1μg, total volume 6μL. Add 1 μL 3’SR Adapter for Illumina by pipetting and mixing, and place in a PCR instrument for 2 minutes at 70°C. After the reaction was completed, 10 μL 3’Ligation ReactionBuf...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com