Primers, detection method and application for diploid Haynaldia villosa 3V chromosome specific KASP marker detection
A technology of marker detection and detection method, which is applied in the field of molecular biology, can solve problems such as time-consuming and labor-intensive, and achieve the effects of improving efficiency, accelerating transfer and utilization, and accelerating the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
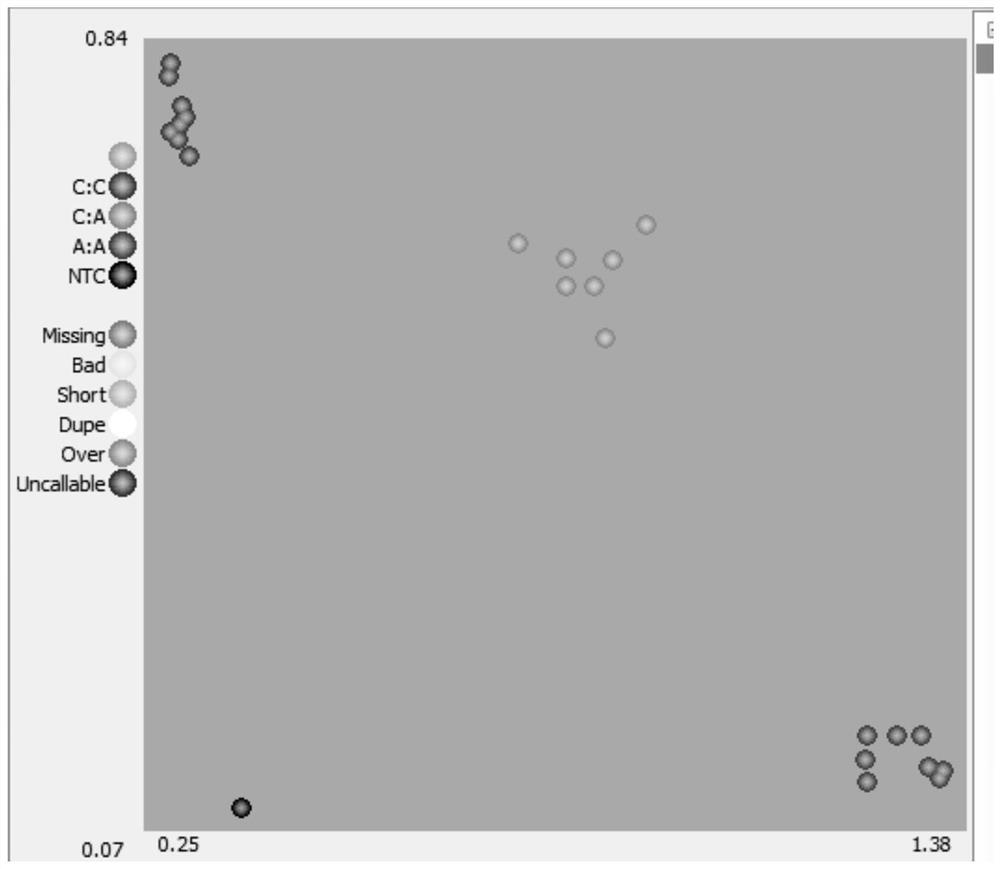
Image
Examples
Embodiment 1
[0041] A preferred embodiment of the present invention provides a diploid 3V chromosome-specific KASP marker detection primer, the nucleotide sequence of the primer is as follows:
[0042] Upstream primer F1: 5'-GAAGGTGACCAAGTTCATGCTGAGCAGGCTGTCGAAGCTA-3';
[0043] Upstream primer F2: 5'-GAAGGTCGGAGTCAACGGATTGAGCAGGCTGTCGAAGCTC-3';
[0044] Downstream primer R: 5'-TGGAGCACGAGGGTGTGA-3'.
Embodiment 2
[0046] The present invention is based on the method for using the above-mentioned primers to detect the 3V chromosome of diploid T. villosa provided by the above-mentioned embodiment 1, comprising the following steps:
[0047] 1. Using the CTAB method to extract the genomic DNA of the plant to be tested, the extraction steps are as follows:
[0048] 1) Take 2g of fresh young leaves, grind them into fine powder with liquid nitrogen, add 2×CTAB extract (2% CTAB; 1.4M NaCl, 0.1M Tris-HCl, pH 8.0, 0.1M EDTA, pH 8.0) 15ml, mix well.
[0049] 2) 30-45min in a water bath at 65°C, during which time shake gently to mix. After cooling to room temperature, add an equal volume of chloroform:isoamyl alcohol (24:1), mix gently until the supernatant becomes milky, and centrifuge at 4000rpm for 10min.
[0050]3) Take the supernatant, add an equal volume of isopropanol, and place in an ice bath to precipitate DNA.
[0051] 4) The DNA was hooked out, washed twice with 70% ethanol and once wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com