Short joint, double-index joint primer and double-index library building system of gene sequencer
A technology of linker primers and linkers, applied in the field of short linkers, can solve the problems of poor uniformity of amplification efficiency, low fragment conversion efficiency, and poor uniformity of capture efficiency among index combinations.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1 short connector
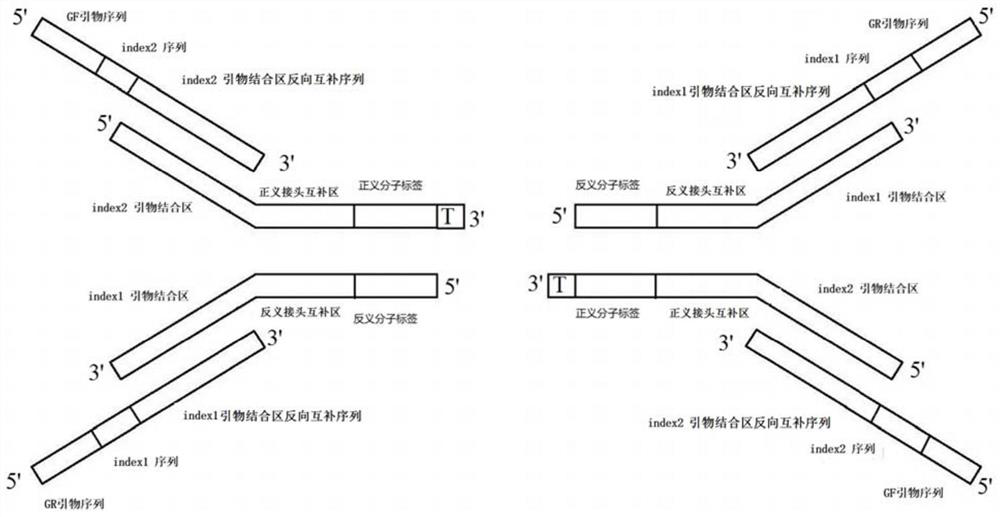
[0044] short connectors, such as figure 1 Shown: the linker oligonucleotide chain 1 includes index2 primer binding region, sense linker complementary region, sense molecular tag and 1 protruding base T from the 5' end to the 3' end, and the sense linker complementary region All or part of it coincides with the 3' end sequence of the index2 primer binding region, and its nucleotide sequence is as follows; the linker oligonucleotide chain 2 includes the linker oligonucleotide chain in turn from the 5' end to the 3' end The antisense molecular label that is reverse complementary to the sense molecular tag in 1, the antisense adapter complementary region that is reverse complementary to the sense adapter complementary region of adapter oligonucleotide chain 1, and the index1 primer binding region, and the antisense adapter complementary region is all Or partially coincide with the 5' end sequence of the index1 primer binding region, and its n...
Embodiment 2
[0059] Example 2 double index linker primer
[0060] Double index linker primers, including forward linker primers and reverse linker primers;
[0061] Forward linker primer, including GF primer sequence, index2 sequence and index2 primer binding region sequence sequentially from 5' end to 3' end;
[0062] Reverse linker primer, including GR primer sequence, index1 sequence, index1 primer binding region reverse complementary sequence from 5' end to 3' end;
[0063] Such as figure 1 Shown: the nucleotide sequence of the forward adapter primer is as follows; the nucleotide sequence of the reverse adapter primer is as follows; specifically as follows:
[0064] Forward linker primer 5'-3':
[0065] TCTCAGTACGTCAGCAGTT NNNNNNNNNNCAACTCCTTGGCTCA CA GAACGACATGGCTACGATCC ;
[0066] Reverse Adapter Primer 5'-3':
[0067] GGCATGGCGACCTTATTCAG NNNNNNNNNNTT ;
[0068] In the above sequence (refer to the sequence table SEQ ID NO.35-36), "NNNNNNNNNN" in the forward adapter p...
Embodiment 3
[0070] Example 3 Double index linker blocking sequence
[0071] Double index adapter blocking sequence: including forward adapter blocking sequence and reverse adapter blocking sequence;
[0072] The forward linker blocking sequence includes the reverse complementary sequence of the sense linker complementary region, the reverse complementary sequence of the index2 primer binding region, the reverse complementary sequence of the index2 sequence and the reverse complementary sequence of the GF primer sequence from the 5' end to the 3' end; All or part of the reverse complementary sequence of the sense linker complementary region coincides with the 5' end sequence of the reverse complementary sequence of the index2 primer binding region;
[0073] Reverse linker blocking sequence, including GR primer sequence, index1 sequence, index1 primer binding region reverse complementary sequence and antisense linker complementary region reverse complementary sequence from 5' end to 3' end;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com