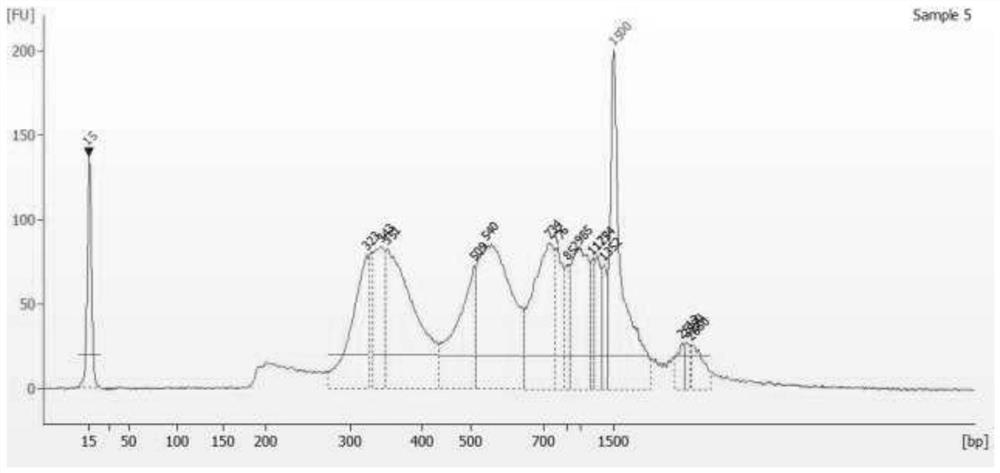
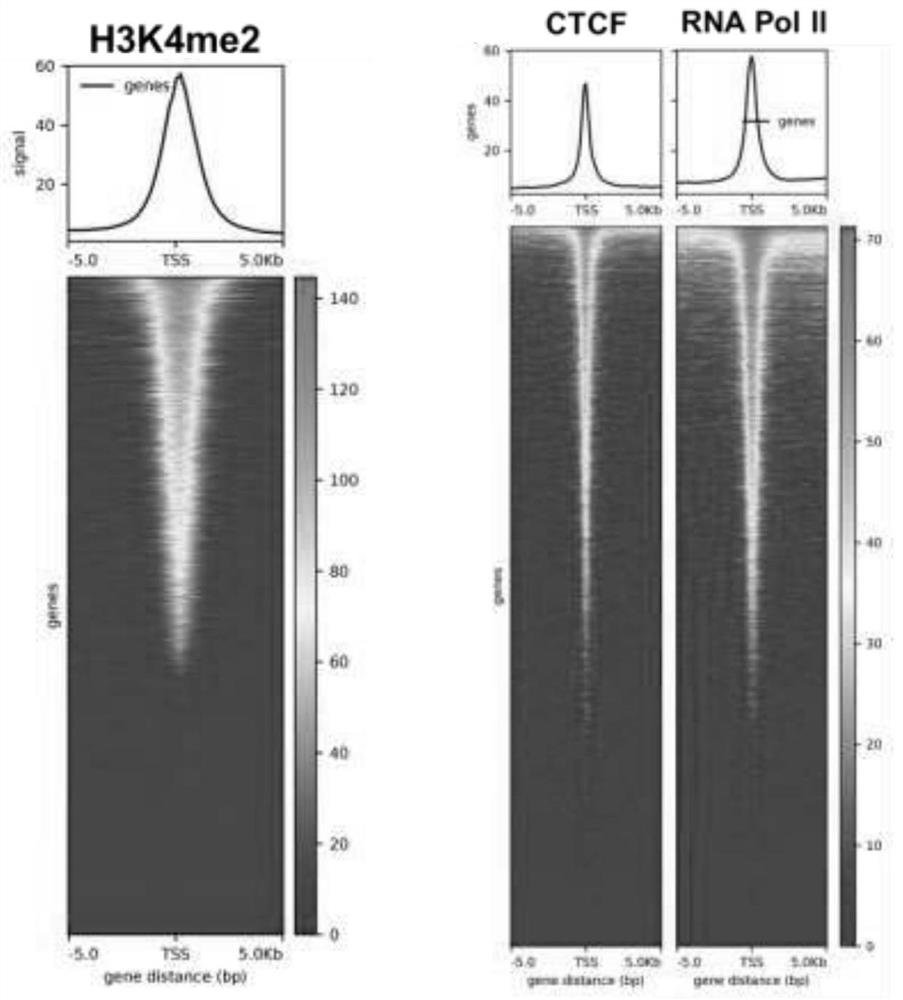
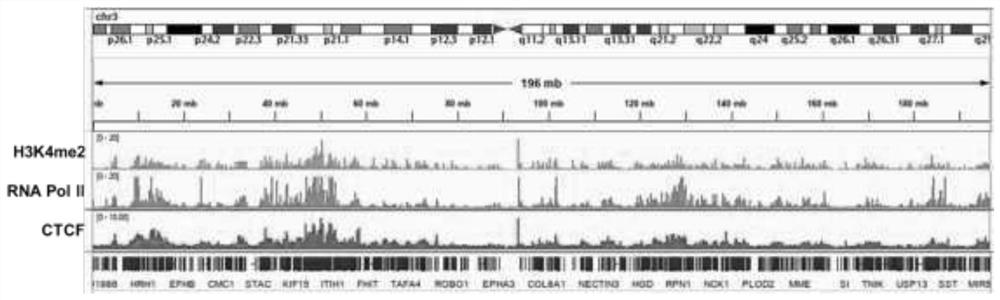
Research method and tool for multi-target protein-DNA interaction
A technology of targeting and targeting molecules, which is applied in the field of targeted transposome complexes to study multi-target protein-DNA interactions, which can solve the problems of false negatives, high background noise of sequencing, and shortened experimental time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0073] An alternative embodiment of the above three oligonucleotide sequences is as follows:
[0074] Oligonucleotide 1' (SEQ ID NO: 9): 5'-phos- AGATGTGTATAAGAGACAG -NH 2 -3'
[0075] Oligonucleotide 2' (SEQ ID NO: 13):
[0076]
[0077] Oligonucleotide 3' (SEQ ID NO: 14):
[0078]
[0079] The above oligonucleotide 2, oligonucleotide 2', oligonucleotide 3 and oligonucleotide 3' constitute yet another alternative embodiment of the present invention.
Embodiment 2
[0080] The index adopted in embodiment 2:
[0081]
Embodiment 3
[0082] Index adopted in embodiment 3:
[0083]
[0084] The Illumina library structure is as follows:
[0085]
[0086] Wherein, -MMMMMM- represents the insertion sequence (the length of the insertion sequence is 6 nucleotides is exemplary, not limiting), and the meanings of other segments are the same as above.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com