Combined marker for early diagnosis of liver cancer and application of combined marker.
A marker, liver cancer technology, applied in the field of biomedicine, can solve the problems of lack of effective therapeutic targets, difficulty in early diagnosis, high recurrence and metastasis rate, avoid tissue biopsy, improve the sensitivity of early screening, and improve the effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Embodiment 1, the establishment of the method for liver cancer detection
[0071] 1. Markers for the detection of liver cancer
[0072] The inventors of the present invention combined various databases and comprehensive clinical information, designed probes to enrich the methylation sites of related genes through large-scale screening, and then compared the cfDNA 3-6 months before the diagnosis of liver cancer by building a library and sequencing The difference in methylation level between samples and cfDNA samples of non-liver cancer patients was used to obtain markers for liver cancer detection. The markers include the degree of methylation of any 6 or more of the following 17 genes and / or the mutation of at least one of the two genes TERT and CTNNB1:
[0073] AK055957, DAB2IP, GRASP, PPFIA1, PSD4, TBX15, MKL1, DCDC2, BCL2L11, SEMA4D, PROX1, FAM78A, PTPN7, AXIN1, SHH, SR, and CYP1B1.
[0074] The positions of these genes (reference genome GRCh38 / hg38) are shown in T...
Embodiment 2
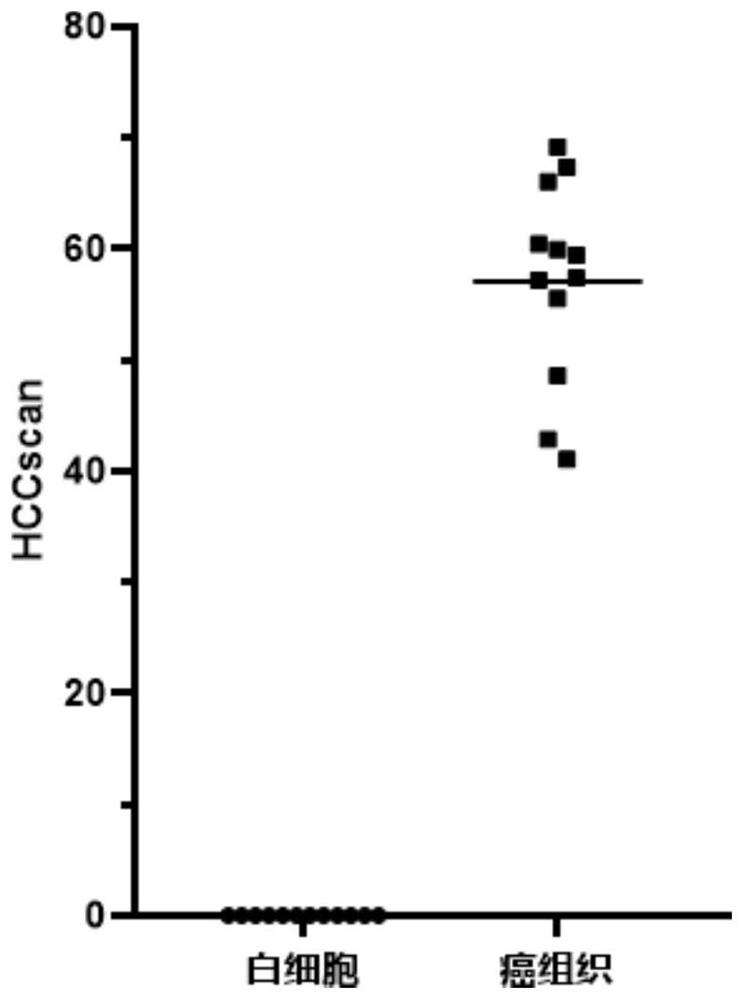
[0125] Example 2, using 7 markers of the present invention to detect samples
[0126] The selected samples were 12 clinical liver cancer tissue samples and 12 healthy white blood cell samples.
[0127] The inclusion criteria for 12 patients with clinical liver cancer were: liver cancer was diagnosed clinically by pathological examination of tissue sections.
[0128] The inclusion criteria of 12 healthy people were: healthy non-liver cancer patients.
[0129] The seven markers are: MKL1, DCDC2, BCL2L11, CYP1B1, PTPN7, AXIN1 and SEMA4D gene methylation.
[0130] 1. The test sample is first passed through Qiagen's Genomic DNA was extracted according to the operation in the instruction manual. The OD260 / 280 of DNA measured by ThermoFisher's Nanodrop should be between 1.8-2.1.
[0131] 2. The extracted DNA is subjected to methylation detection, according to the method of step 2 of Example 1, transformation, pre-amplification and final qPCR detection are carried out, and the re...
Embodiment 3
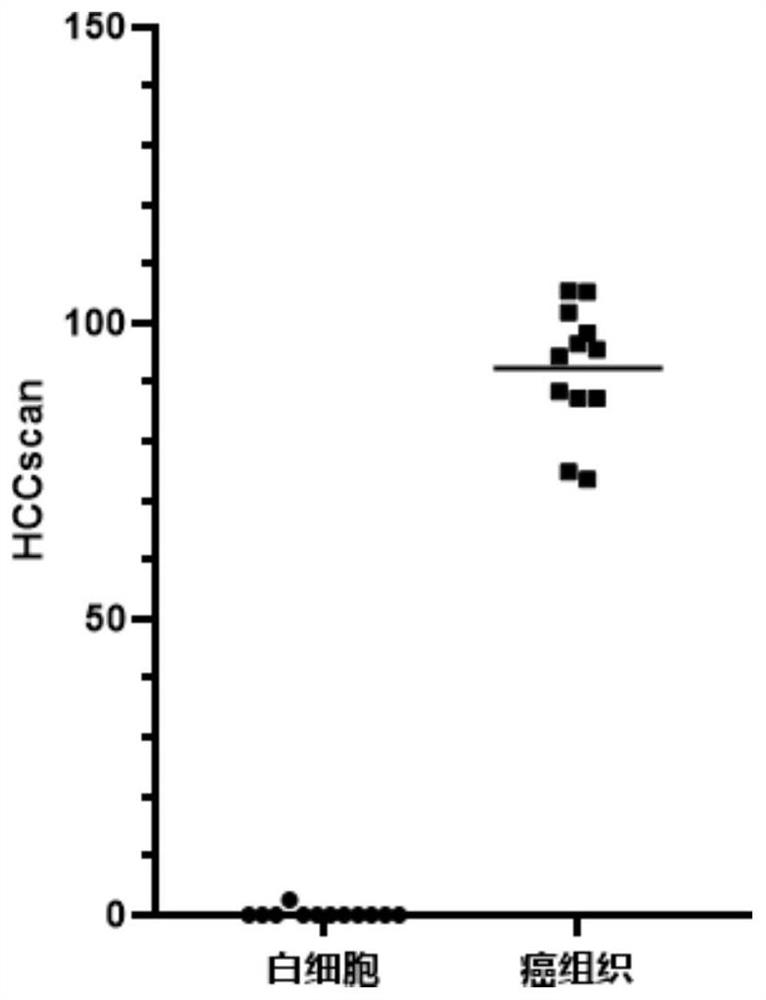
[0137] Example 3, using 10 markers of the present invention to detect samples
[0138] Selected sample is with embodiment 2.
[0139] 10 markers: FAM78A, MKL1, DCDC2, BCL2L11, PTPN7, AXIN1, PROX1, AK055957, PPFIA1 and PSD4 gene methylation.
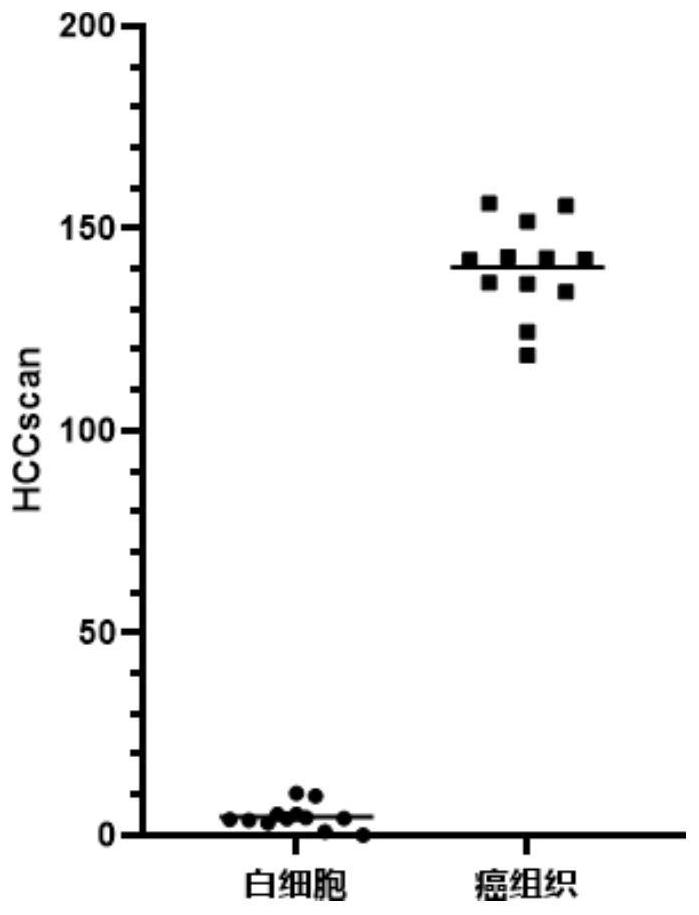
[0140] The detection steps are the same as in Example 2, and the methylation of ten genes FAM78A, MKL1, DCDC2, BCL2L11, PTPN7, AXIN1, PROX1, AK055957, PPFIA1, and PSD4 are detected according to the methylation detection method in Step 2 of Example 1 , and calculate the normalized value L of these ten genes respectively 目的基因 , the normalized value L of these ten genes 目的基因 Sum to get HCCscan, HCCscan=L FAM78A +L MKL1 +L DCDC2 +L BCL2L11 +L PTPN7 +L AXIN1 +L PROX1 +L AK055957 +L PPFIA1 +L PSD4 , the results of each sample are shown in Table 11.
[0141] Table 11. Test results
[0142] sample name sample type HCCscan sample name sample type HCCscan T25 cancer tissue 105.40 WBC10 leukocyte 0.00 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com