Construction of broad-host plasmid for inducible expression of green fluorescent protein and application of broad-host plasmid in fluorescent tracing
A green fluorescent protein and induced expression technology, applied in the field of genetic engineering, can solve the problems of degradation, inability to use, incompatibility, etc., achieve strong fluorescence activity, eliminate interference from dead bacteria, and facilitate observation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
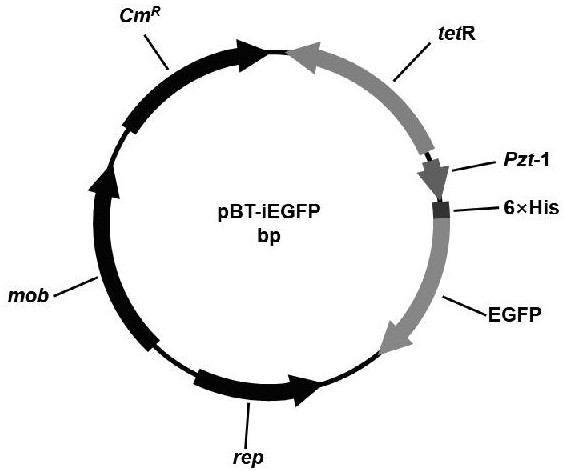
[0038] Example 1 Construction of pBT-iEGFP plasmid
[0039] The inducible plasmid pZT-EGFP plasmid (Tian M, Qu J, Bao Y, Gao J, Liu J, Wang S, Sun Y, Ding C, Yu S. Construction of pTM series plasmids forgene expression in Brucella species. J Microbiol Methods, 2016; 123: 18-23.) as a template, iEGFP-F and iEGFP-R as primers, the primers are as follows:
[0040] iEGFP-F: AGGGAACAAAAGCTGGGTACCCCGTTTCCATTTAGGTGGGTA (SEQ ID NO. 2)
[0041] iEGFP-R: CGCGGTGGCGGCCGCTCGATCGATTATTTTATTTCCTG (SEQ ID NO. 3)
[0042] Use the fidelity enzyme PrimeSTAR Max DNA Polymerase (Dalian Bao Biological Co., Ltd.) to perform PCR amplification of the gene fragment TetR-Pzt-1-iEGFP that can induce the expression of green fluorescent protein. The PCR reaction system is 50 µL, including: PrimeSTAR Max Premix (2× ) 25 µL, each 2 µL of upstream primer and downstream primer, 2 µL of pZT-EGFP (5 µg / µL), make up to 50 µL with sterile water; 30 s for 30 cycles. The size of the product was 1908 bp. After...
Embodiment 2
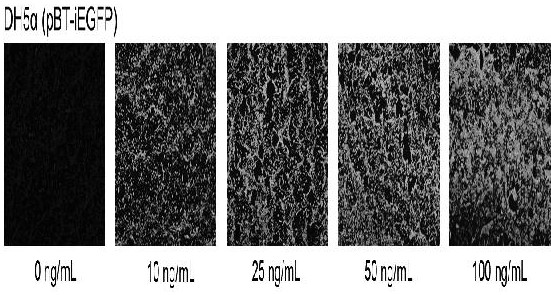
[0045] Example 2 Induced expression of green fluorescent protein by pBT-iEGFP plasmid in Escherichia coli
[0046] Induced expression of green fluorescent protein by pBT-iEGFP plasmid in Escherichia coli engineering strain DH5a
[0047] Take 100 µL Escherichia coli competent cells (Beijing Tiangen Biology), slowly melt on the ice-water mixture, add 20ng pBT-iEGFP plasmid, mix well, ice bath for 30 min, heat shock at 42°C for 90 s, and ice bath for 2 min , add 1 mL of LB solution, shake and incubate at 37 °C for 45 min, spread chloramphenicol on LB plates to screen positive clones, identify positive colonies by PCR, and name the positive bacteria as DH5α(pBT-iEGFP); inoculate DH5α(pBT-iEGFP) on Add 0 ng / mL, 10 ng / mL, 25 ng / mL, 50 ng / mL, and 100 ng / mL anhydrotetracycline to the chloramphenicol LB solution to induce expression for 12 h, take 500 µL of culture solution, and wash twice with PBS. Suspend in sterile water, take 5-10 µL of the suspension to coat glass slides, dry wit...
Embodiment 3
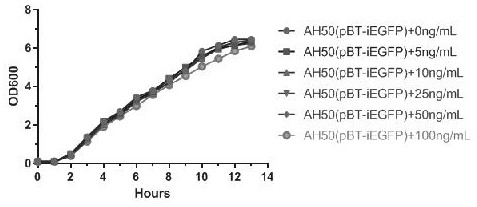
[0053] Example 3 pBT-iEGFP plasmid induces expression of green fluorescent protein in Salmonella typhimurium
[0054] 1. Construction of recombinant bacteria: Salmonella typhimurium SL1344 was inoculated into LB liquid medium, cultured to early logarithmic growth (OD600= 0.4-0.6), ice bathed for 10 min, washed with sterile pre-cooled water twice, Pre-cooled 10% glycerol sterile water to suspend cells, aliquot 100 µL in each tube, add 1 µg pBT-iEGFP plasmid, ice-bath for 10 min, add the mixture to a 1 mm electroporation cuvette, under the condition of 1.8 kV 200 Ω, The plasmid was electrotransformed into Salmonella typhimurium SL1344, chloramphenicol was applied on LB plates to screen positive clones, and PCR was used to identify positive clones. The identified positive strain was named SL1344 (pBT-iEGFP).
[0055] 2. Recombinant bacteria are sensitive to anhydrotetracycline: SL1344(pBT-iEGFP) was inoculated in LB liquid medium, cultured for 12 hours, measured the OD600 value o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com