Biological sequencing
A biological sequence and sequencing technology, applied in bioinformatics, sequence analysis, instruments, etc., can solve problems such as missing, error, and enclosure, and achieve the effects of reducing errors, increasing speed, and reducing complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0182] Example 1: Sequencing according to an embodiment of the invention
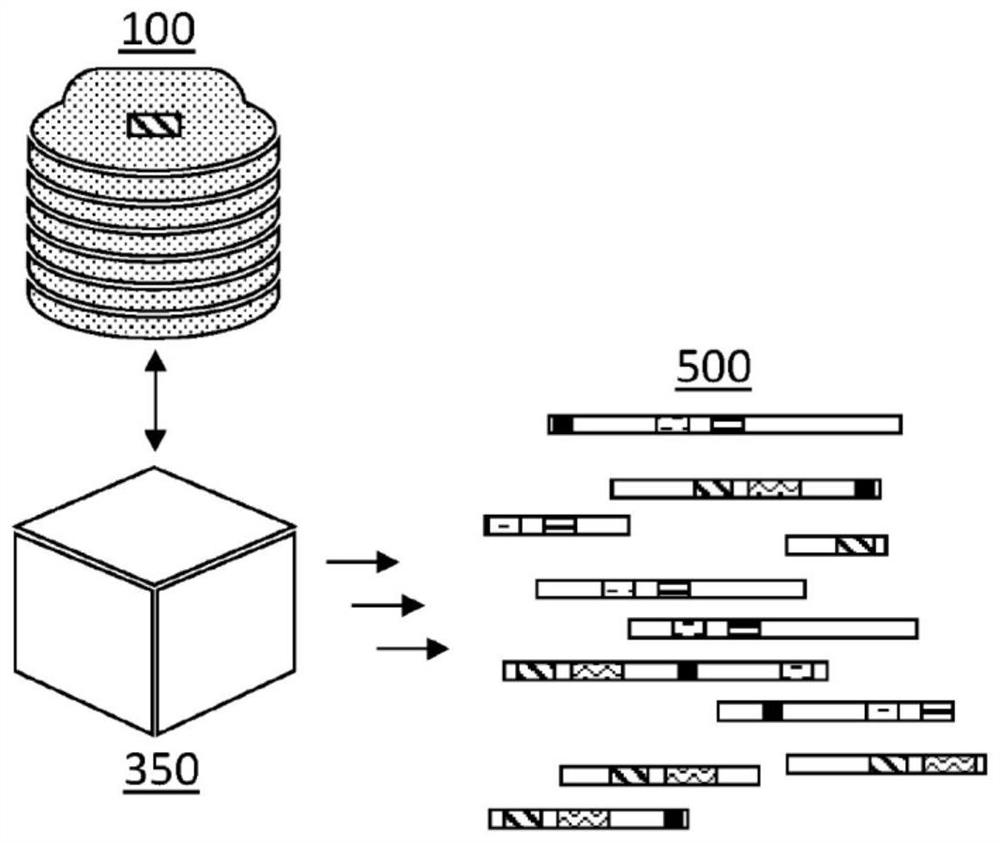
[0183] By way of illustration, embodiments of the present invention are not limited in this regard, Figure 7 Examples of possible sequencing implementations are shown in . The figures illustrate possible different method steps of a sequencing method according to an embodiment of the invention. The method comprises, after obtaining at least a first read of a biopolymer or biopolymer fragment, and typically during the further receipt of reads of a biopolymer or biopolymer fragment to be sequenced, parsing incoming, e.g., received , reads with fingerprints, called HYFTs TM . After resolution, an alignment (eg, matching) can be performed in order to obtain a map representing the sequence of the biopolymer or biopolymer fragment. Alignment can be performed by alignment with directed graphs, such as directed acyclic graphs. The latter may be a universal genome reference map, but embodiments are not li...
Embodiment 2
[0184] Embodiment 2: the processing of protein database
[0185] Example 2a: About HYFT found in the protein database TM Analysis of fingerprints against protein databases
[0186] To illustrate HYFT TM The ubiquity of fingerprints in biological sequence databases, taking the Protein Data Bank (PDB) as an example of a large, commonly available biological sequence database, and using a repository of fingerprint data strings obtained as described above was processed according to the invention. The results were analyzed with respect to various indicators, a selection of which is given below.
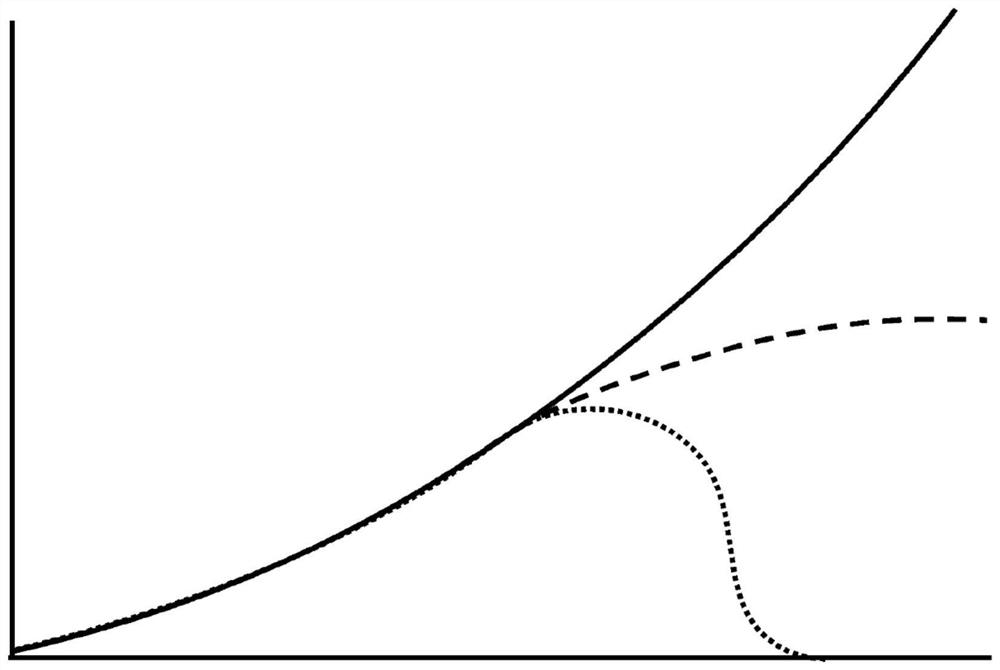
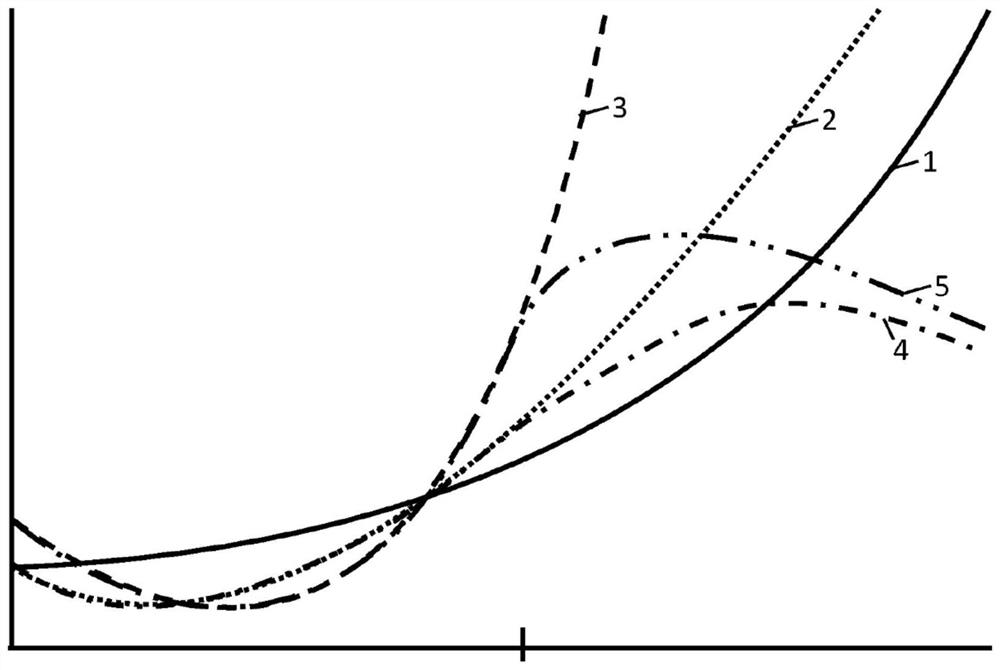
[0187] Figure 12 and Figure 13 HYFT of processed protein sequences of lengths up to 50 and lengths up to 5000+ are shown, respectively TM Coverage (in %). Here, coverage is the sequence unit in the total sequence length attributed to HYFT TM part of the fingerprint. In other words, coverage is the combined length of one or more first parts divided by the total sequence length. ...
Embodiment 3
[0197] Example 3: Comparison between sequence searches known in the prior art and those described herein
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com