Primer combination for detecting SARS-CoV-2 and D614G mutant strain thereof and application of primer combination
A technology of D614G and primer combination, applied in the field of biomedicine, can solve the problems of unfavorable case screening, high sample requirements, limited sample capacity, etc., and achieve the effect of reducing false negative results, good application prospects, and low detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
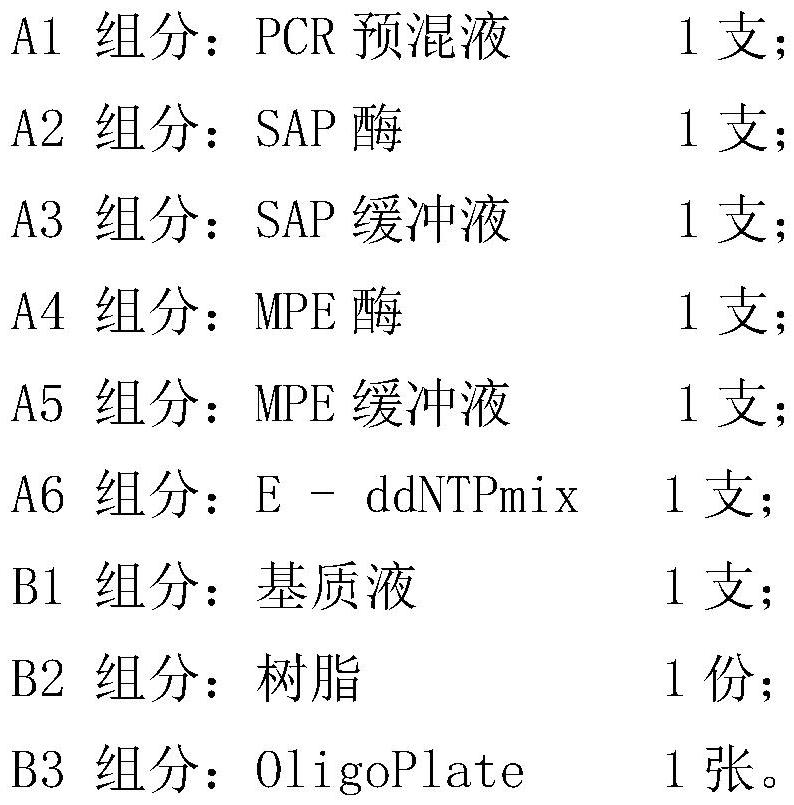
[0119] Embodiment 1, the preparation of the primer combination that detects novel coronavirus
[0120] 1. Preparation of primer combinations
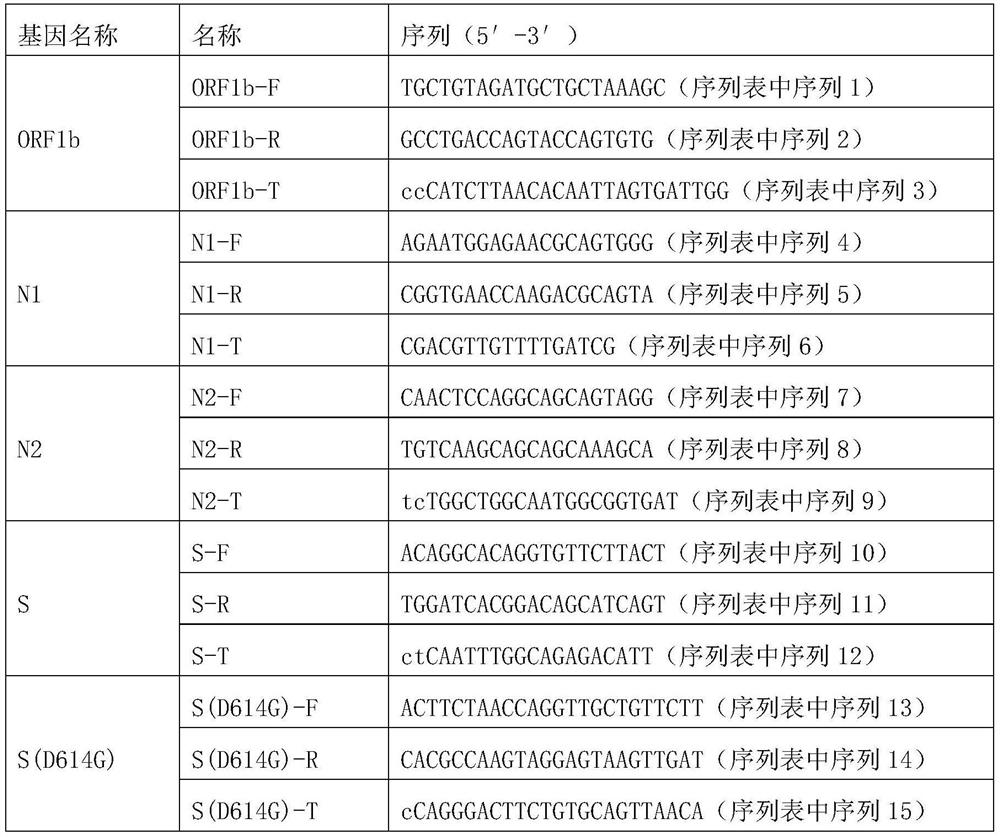
[0121] In this example, a primer combination for the detection of novel coronavirus was prepared. MPE primer composition for T, N1-T, N2-T, S-T and S(D614G)-T. ORF1b-P is composed of single-stranded DNA named ORF1b-F and ORF1b-R, and its sequences are sequence 1 and 2, respectively; N1-P is composed of single-stranded DNA named N1-F and N1-R, respectively. The sequences are sequence 4 and 5 respectively; N2-P is composed of single-stranded DNA named N2-F and N2-R respectively, and its sequence is sequence 7 and 8 respectively; S-P is composed of single-stranded DNA named S-F and S-R respectively , whose sequences are sequences 10 and 11, respectively; S(D614G)-P is composed of single-stranded DNA named S(D614G)-F and S(D614G)-R, respectively, and whose sequences are sequences 13 and 14, respectively; ORF1b- The sequences of T, N1-T, N2...
Embodiment 2
[0141] Embodiment 2. Establishment of a method for detecting novel coronaviruses using the primer combination of Embodiment 1
[0142] Detect the amplification efficiency of the primer combination of embodiment 1, adopt SNP typing kit (QT-SJ09-SNPs) to carry out, the steps are as follows:
[0143] 1. Prepare the primer mixture
[0144] Preparation of primer mixture: use RNase-free deionized water to dissolve the primer pairs ORF1b-P, N1-P, N2-P, S-P and S(D614G)-P in Example 1 to obtain a primer mixture. In the primer mixture , the concentrations of ORF1b-F, ORF1b-R, N1-F, N1-R, N2-F, N2-R, S-F, S-R, S(D614G)-F and S(D614G)-R were all 0.5 μM.
[0145] 2. Multiplex PCR amplification
[0146] Use the primer mixture in step 1 to carry out multiplex PCR amplification, the DNA template is pUC57-SARS-CoV-2 in Example 1, and the system is as follows:
[0147] components volume / μl PCR master mix 2 Deionized water 1 primer mix 1 DNA template (at a co...
Embodiment 3
[0186] The specificity of the primer combination of embodiment 3, embodiment 1
[0187] pUC57-SARS-CoV-2, pUC57-HCoV-229E, pUC57-HCoV-OC43, pUC57-HCoV-NL63, pUC57-HCoV-HKU1-1, pUC57-HCoV-HKU1-2, pUC57-SARS of Example 1 –CoV and pUC57-MERS-CoV utilize RNase-free ddH respectively 2 O was dissolved, and the concentration of the plasmid obtained was 10 5 Eight plasmid solutions in copies / μl, namely pUC57-SARS-CoV-2 solution, pUC57-HCoV-229E solution, pUC57-HCoV-OC43 solution, pUC57-HCoV-NL63 solution, pUC57-HCoV-HKU1–1 solution, pUC57 -HCoV-HKU1–2 solution, pUC57-SARS–CoV solution, and pUC57-MERS-CoV solution.
[0188] Using the method 2-6 in Example 2, the DNA templates were respectively replaced with the above-mentioned eight kinds of plasmid solutions, and the other steps were kept unchanged, and the specificity of the primer combination in Example 1 was tested.
[0189] The results are shown in Table 4. The specificity of the primer combination in Example 1 is very good, th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com