mTORC1 INHIBITORS
A technology of inhibitors and excipients, applied in the directions of antitoxins, anti-inflammatory agents, antiviral agents, etc., can solve problems such as Akt activation and blocking
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
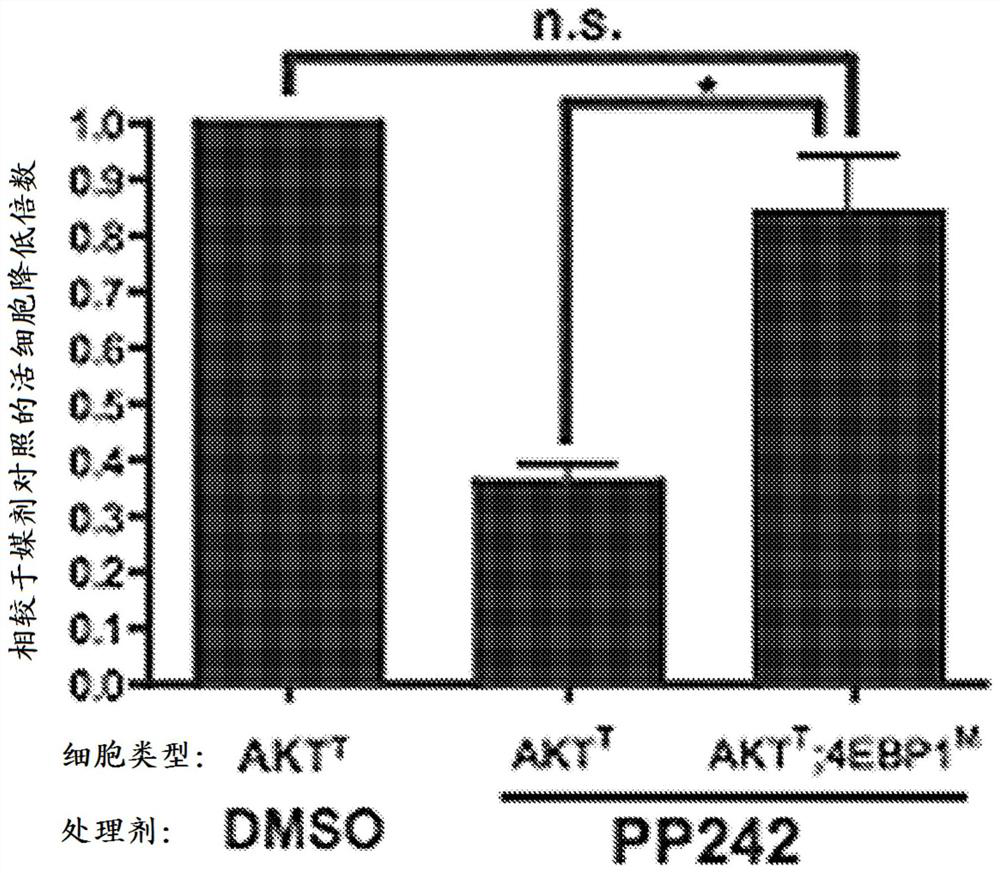
[0374] The effect of MLN0128 on TORC2 is not considered to be necessary for its anticancer activity. This has been analyzed in a genetically engineered mouse model of Akt-driven lymphomagenesis 10 . A nonphosphorylatable mutant of 4E-BP1 that constitutively inhibits eIF4E blocks Akt-driven lymphomagenesis, thereby indicating that this single substrate of mTOR is sufficient to support cancer cell growth and survival. In some animals with constitutively active Akt and constitutively active 4E-BP1, some tumors still express 4E-BP1 mutants (4EBP1 M ). Presumably, bypassed by 4EBP1 M Another pathway of applied blockade has been activated. These cells provide genetic testing of the mechanism of action of inhibitors of the asTORi class. If the effects of asTORi on cells are mediated by 4E-BP1 phosphorylation, then when added to bypass 4EBP1 M The drug should have no effect when in the cells. asTORi PP242 had no significant effect in the cells ( figure 1 ), thus confirming th...
Embodiment 2
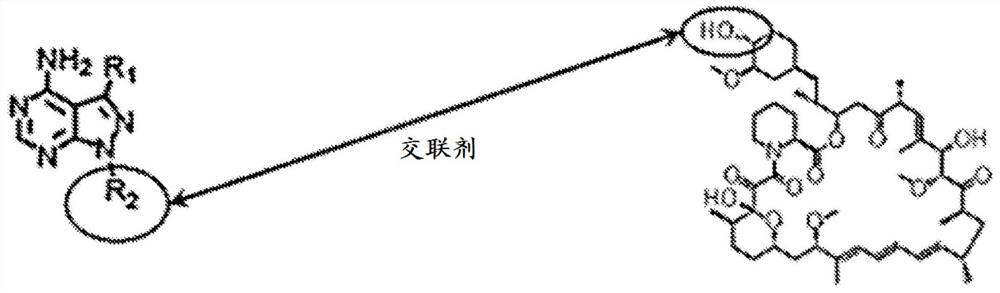
[0378] Design and production of a third class of mTOR inhibitors - "rapamycin-linkers". In the last year, part of the crystal structure of mTOR was solved by Pavletich and colleagues 17 . This structure corresponds to the much earlier structure of rapamycin binding to the isolated FRB and FKBP proteins 18 The combination offers the opportunity to develop a completely new class of mTOR inhibitors. By exploiting the TORC1-selective properties of rapamycin, and linking rapamycin to (in a manner that does not disrupt rapamycin binding to the FKBP12 or the FRB domain of mTOR) asTORi (eg, MLN0128), we predict that the development of A new class of pharmacological agents for targeting mTOR ( Figure 2A and 2B ). This method to design mTOR inhibitors has never been proposed before and provides a novel pharmacological means of blocking mTOR signaling. A key aspect of this design is the use of rapamycin to "deliver" asTORi (eg, MLN0128) into the active site of mTORC1, effectively ...
Embodiment 3
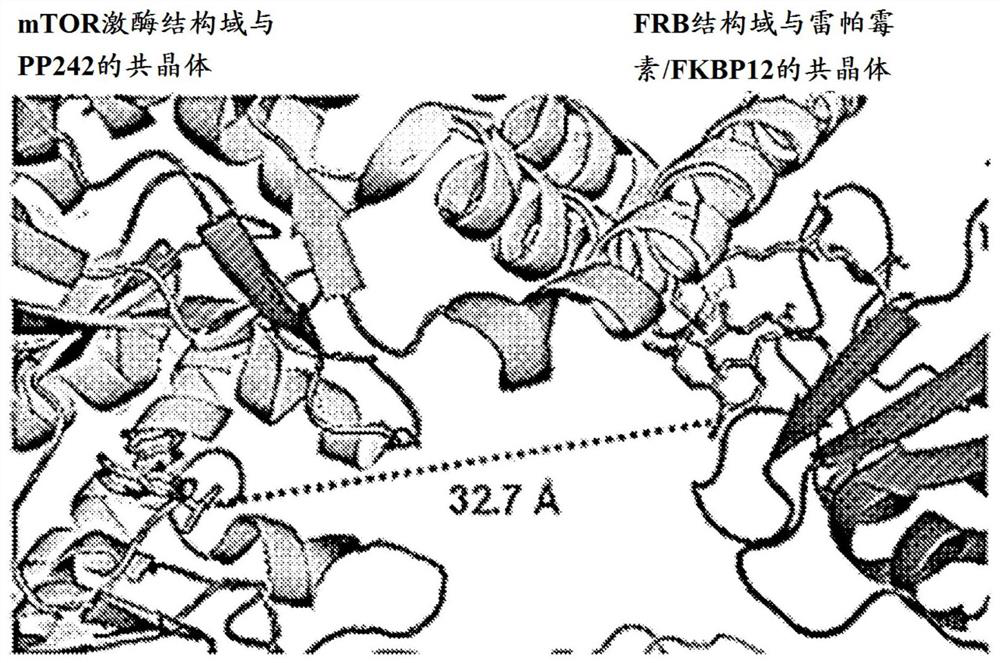
[0381] To determine the optimal regiochemistry and crosslinker length for tethering rapamycin to asTORi (e.g. MLN0128), mTOR (PDB:4JT5) was stacked using the consensus FRB domain contained in both structures 17 and FRB:FKBP12:rapamycin (1FAP) 18 The two independent crystal structures of ( Figure 4A ).
[0382] To design the hybrid compounds, the tethering sites for rapamycin were designed in a manner that minimized disruption of rapamycin binding to the FKBP12 protein and the FRB domain of mTOR. Similarly, the linking group of asTORi (eg, MLN0128) is designed to avoid disrupting the binding of the active site inhibitor to the catalytic domain of mTOR. Co-crystal structure of rapamycin (1FAP) 18 Analysis of rapamycin revealed that the hydroxyl group at the C40 position of rapamycin is exposed to the solvent region and is oriented towards the active site of mTOR ( Figure 4A ). For asTORi (e.g. MLN0128), tethering sites were selected based on the co-crystal structure of mT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com