L-phenylalanine producing strain and construction method thereof
A technology of phenylalanine and construction method, which is applied in the field of genetic engineering, can solve the problems of no acid production, OD value drop, etc., and achieve the effects of increasing production, solving OD drop, and increasing metabolic flow
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: Codon optimization
[0042] The nucleotide sequence of the pheA gene obtained by the inventor from the existing database is shown in SEQ ID NO.1, and the amino acid sequence of the encoded chorismate mutase is shown in SEQ ID NO.2. The nucleotide sequence of the aroF gene is shown in SEQ ID NO.3. The nucleotide sequence of the aroB gene is shown in SEQ ID NO.4. The nucleotide sequence of the aroE gene is shown in SEQ ID NO.5.
[0043] In order to make the pheA gene, aroF, aroB gene and aroE gene better adapt to the prokaryotic expression system, the present invention optimizes the codons of the above genes respectively.
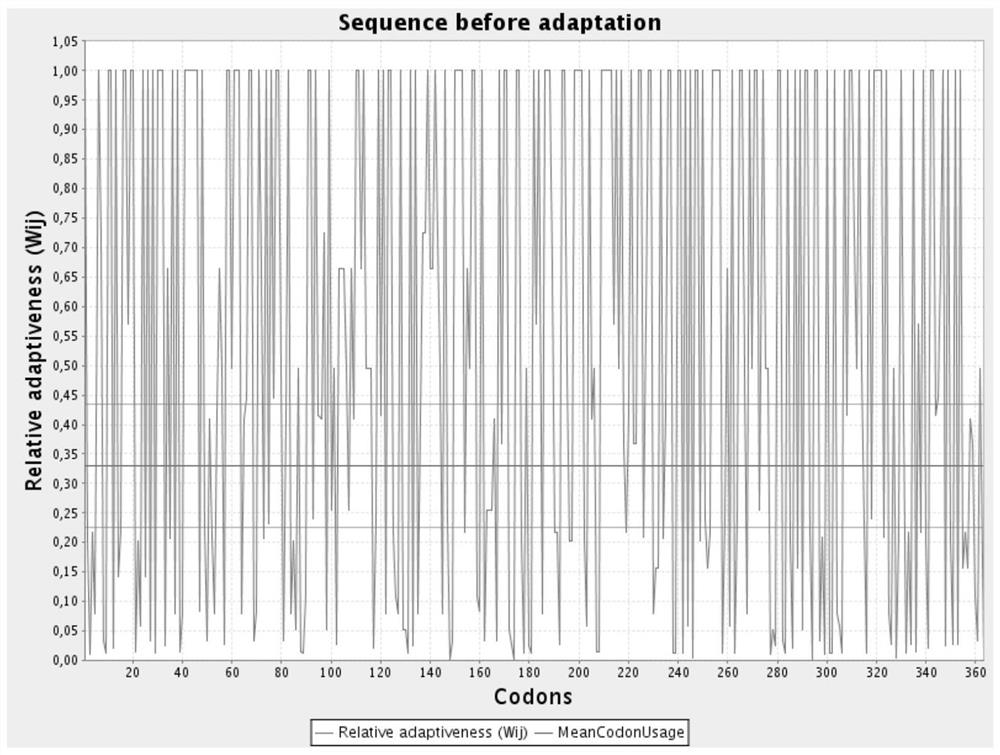
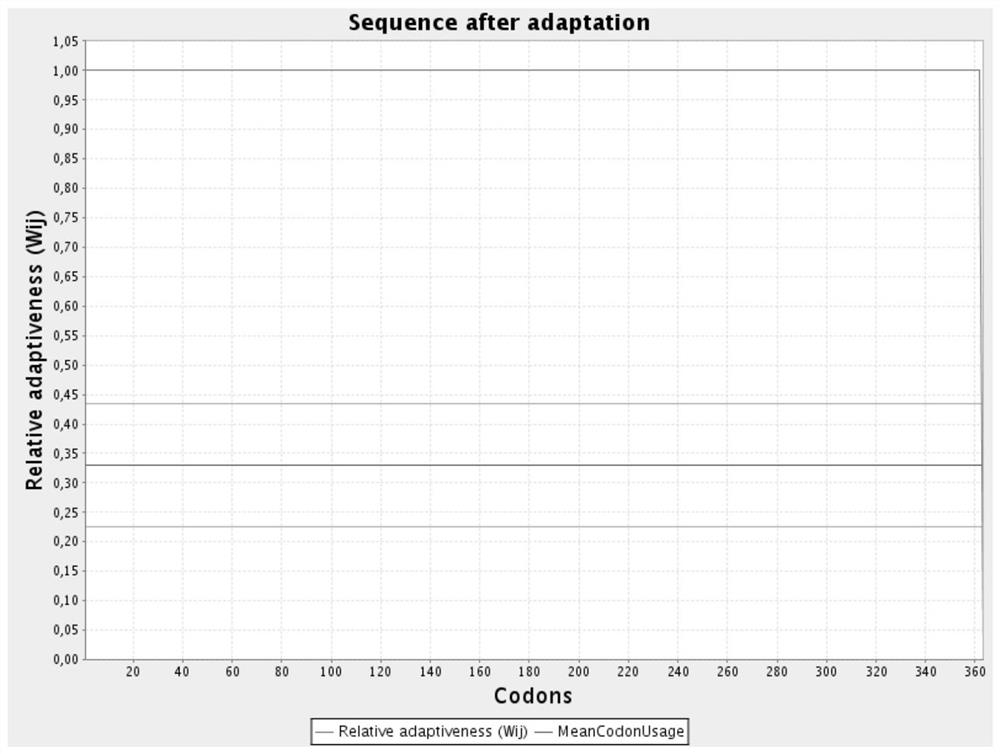
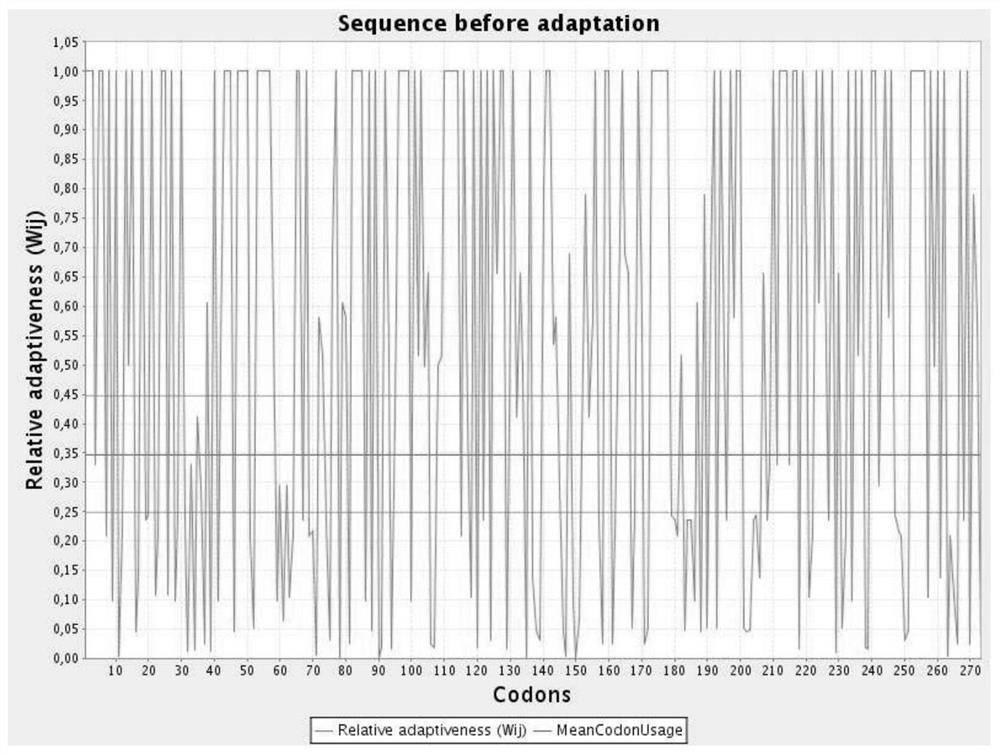
[0044] The nucleotide sequence of the aroB gene after codon optimization is shown in SEQ ID NO.6; the codon relative fitness figure before optimization is shown in figure 1 Shown; the codon relative fitness map after optimization is shown in figure 2 shown.
[0045] The nucleotide sequence of the aroE gene after codon optimization is s...
Embodiment 2
[0048] Embodiment 2: Construction of the first recombinant expression vector
[0049] Plasmid pET-28a(+) was digested with NcoI and SacI, and then the aroB gene (shown in SEQ ID NO.6) was integrated into the double-digested plasmid pET-28a(+) to obtain a recombinant plasmid pET-aroB; then digest the recombinant plasmid pET-aroB with EagI and XhoI, and then integrate the aroE gene (shown in SEQ ID NO.7) into the digested recombinant plasmid pET-aroB to obtain the first Recombinant expression vector (pET-aroB-aroE).
[0050] The constructed first recombinant expression vector was digested with four enzymes NaoI, SacI, EagI and XhoI, and the results were as follows: Figure 9 shown. The results showed that the aroB gene (shown in SEQ ID NO.6) and aroE gene (shown in SEQ ID NO.7) had been successfully integrated into the plasmid pET-28a(+).
Embodiment 3
[0051] Embodiment 3: Construction of the second recombinant expression vector
[0052] Plasmid pGEX-2T was double-digested with BamHI and EcoRI, and then the aroF gene (shown in SEQ ID NO.8) was integrated into the double-digested plasmid pGEX-2T to obtain recombinant plasmid pGEX-2T-aroF, The recombinant plasmid pGEX-2T-aroF was digested with TthllllⅠ and AatⅡ, and the pheA gene (shown in SEQ ID NO.10) was integrated into the digested recombinant plasmid pGEX-2T-aroF to obtain the second Recombinant expression vector (pGEX-2T-aroF-PheA).
[0053] The constructed second recombinant expression vector was digested with four enzymes TthllllⅠ, AatⅡ, BamHI and EcoRI for verification, the results are as follows: Figure 10 shown. The results showed that the aroF gene (shown in SEQ ID NO.8) and pheA gene (shown in SEQ ID NO.10) had been successfully integrated into the plasmid pGEX-2T.
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com