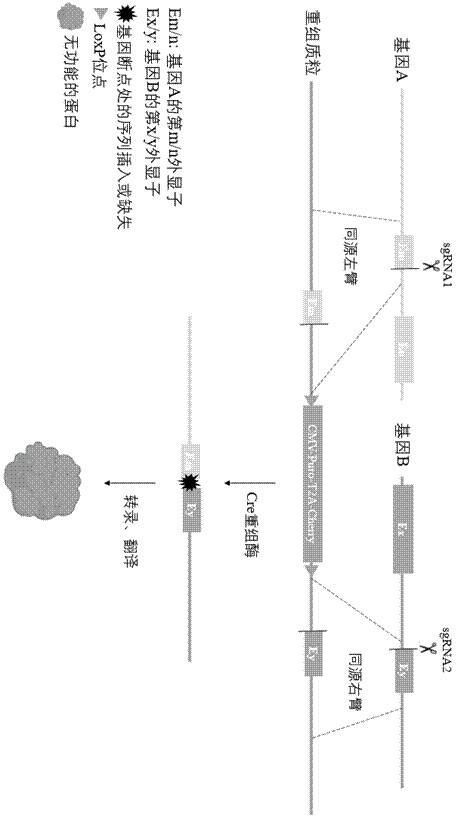
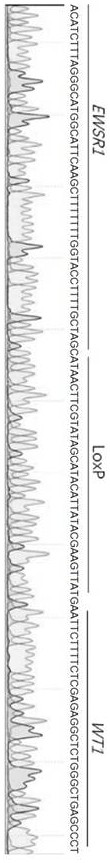
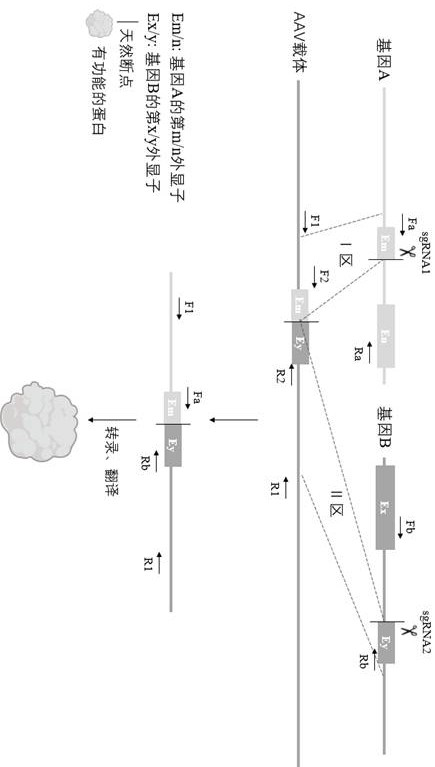
Method for editing gene fusion
An editing and gene technology, applied in the molecular field, can solve the problems of termination, unfavorable molecular diagnostic detection performance, open reading frame destruction of translation, etc., to achieve the effect of improving the success rate and overcoming the inaccuracy of breakpoint sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1 Editing of fusion gene COSF571
[0054] COSF571 is ETV6 Gene (ENST00000396373.8) exon 5 and NTRK3 For the fusion between the 15th exon of the gene (ENST00000394480.6), the sequences of 200 bp upstream and downstream of the natural breakpoint were respectively selected for display. The sequence is shown in SEQ ID NO.1, and the details are as follows:
[0055] CAGCCCCATCATGCACCCTCTGATCCTGAACCCCCGGCACTCCGTGGATTTCAAACAGTCCAGGCTCT CCGAGGACGGGCTGCATAGGGAAGGGAAGCCCATCAAACCTCTCTCATCGGGAAGACCTGGCTTACATGAACCACAT CATGGTCTCTGTCTCCCCGCCTGAAGAGCACGCCATGCCCATTGGGAGAATAGCAG / / ATGTGCAGCACATTAAGA GGAGAGACATCGTGCTGAAGCGAGAACTGGGTGAGGGAGCCTTTGGAAAGGTCTTCCTGGCCGAGTGCTACAACCT CAGCCCGACCAAGGACAAGATGCTTGTGGCTGTGAAGgtaaaccccagaggcatgccggcaccaggagggagggctg gctgagggcccagggagggaaggaagaggc
[0056] Among them, the italic part is ETV6 gene sequence; the underlined part is NTRK3 Gene sequence, " / / " is the natural breakpoint position of the fusion gene, NTRK3 Capital ...
Embodiment 2
[0081] Example 2 Editing of fusion gene ROR1-DNAJC6
[0082] ROR1-DNAJC6 is ROR1 Partial region of exon 9 of gene (ENST00000371079.6) and DNAJC6 For the fusion between the first intron of the gene (ENST00000371069.5), the sequences of 200 bp upstream and downstream of the natural breakpoint were respectively selected for display. The sequence is shown in SEQ ID NO.14, and the details are as follows:
[0083] TCTCTTCTGATTCAGATATCTGGTCCTTTGGGGTTGTCTTGTGGGAGATTTTCAGTTTTGGACTCCAG CCATATTATGGATTCAGTAACCAGGAAGTGATTGAGATGGTGAGAAAACGGCAGCTCTTACCATGCTCTGAAGACT GCCCACCCAGAATGTACAGCCTCATGACAGAGTGCTGGAATGAGATTCCTTCTAGG / / aaacaacttcccaaggtt gtacagcgattacagggtgaagctgccacctgtttatcttagtgcactggagcctgaggtcattgcccactcagtt tgatgccagacagaccagagttgctcattaacacttcataaagccctctcatgttcctcataataaccagacattt ctaattataacttgagcagcaaatgttgtt
[0084] The italic part of the sequence is ROR1 Gene sequence, the underlined part is DNAJC6 Gene sequence, " / / " is the natural breakpoint position...
Embodiment 3
[0098] Example 3 Editing of fusion gene COSF1347
[0099] COSF1347 is FGFR3 Gene (ENST00000440486.8) intron 17 and BAIAP2L1 For the fusion between the first intron of the gene (ENST00000005260.9), the sequences of 200 bp upstream and downstream of the natural breakpoint were respectively selected for display. The sequence is shown in SEQ ID NO.19, and the details are as follows:
[0100] CTTCAAGCAGCTGGTGGAGGACCTGGACCGTGTCCTTACCGTGACGTCCACCGACgtgagtgctggct ctggcctggtgccacccgcctatgcccctccccctgccgtccccggccatcctgccccccagagtgctgaggtgtg gggcgggccttctggggcacagcctgggcacagaggtggctgtgcgaagaggggct / / ctttccacctcggggttc agaaggggactttacgcgggaaggtactttccctccctccagctcccctcccccgcgtccttccacctctcccggt ctctcccactcctcccctggccctccacagcccctcttcttcctcccctggccctctccttcctcccagtccctcc ccatcccctcccccctacttttcctcctcc
[0101] The italic part of the sequence is FGFR3 Gene sequence, the underlined part is BAIAP2L1 Gene sequence, " / / " is the natural breakpoint position of the fusion gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com