Gene for osteosarcoma typing and osteosarcoma prognosis evaluation and application thereof
An osteosarcoma and genetic technology, applied in the field of biomedicine, can solve the problems of inability to well explain patient heterogeneity, inability to well guide patients in personalized treatment such as targeted therapy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049]Example 1 Gene Screening for Osteosarcoma Type and Estimation of Osteosarcoma Prognosis
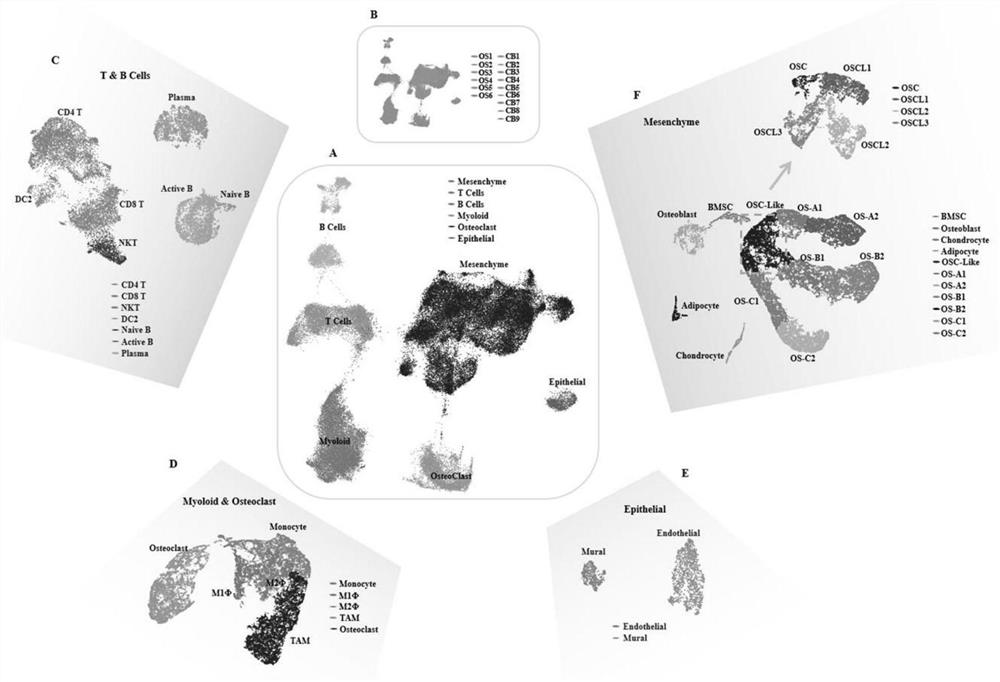
[0050] Through the screening of single-cell high-throughput sequencing data in the GEO public database (the full name is Gene Expression Omnibus, which is a gene expression database created and maintained by the National Center for Biotechnology Information NCBI), we screened out an osteosarcoma numbered GSE152048 10x single-cell sequencing data, 6 cases of classical osteosarcoma (osteogenic osteosarcoma) data were selected for subsequent analysis. In addition, single-cell sequencing data of cancellous bone samples from 9 degenerated patients undergoing surgery in Shanghai Changzheng Hospital were selected. The above two data sets were combined for molecular classification and prognosis analysis of osteosarcoma.
[0051] Visualized UMAPs and different cell clusters were obtained by unsupervised dimensionality reduction and cluster analysis on the above datasets. UMAP (Uniform Mani...
Embodiment 2
[0069] Example 2 TARGET-OS classification and survival analysis
[0070] Validation in the overall osteosarcoma public database samples, osteosarcoma sequencing data were screened from The Cancer Genome Atlas (TCGA) database. Normalized RNA-Seq FPKM and Clinic files were downloaded from the TCGA Data Portal on January 30, 2021.
[0071] According to the expression levels of 44 genes in each case sample in the Target-OS dataset, 85 osteosarcoma cases and clinical information were classified, and we can divide the clinical cases in the Target-OS dataset into corresponding Cluster1, Cluster2 and Cluster3. For example, patients with high expression of type A genes and low expression of type B and C genes are defined as Cluster1, patients with high expression of type C genes and low expression of type B and A genes are defined as Cluster2, patients with high expression of type B genes, Patients with low expression of type A and type C genes were defined as Cluster3. We show the ...
Embodiment 3
[0073] Embodiment 3 analytical method
[0074] The single-cell sample preparation, sequencing and analysis methods described in Example 1 are as follows:
[0075] 1. Sample preparation and sequencing
[0076] All samples were cold-chain transported to the laboratory within 1 hour of separation. Following standard 10x Genomics sample preparation, tissue samples were first cut into 2-4 mm sized pieces, digested with collagenase, and incubated in a shaker at 37°C. The digested and incubated cell suspension is filtered through a mesh strainer, and the filtered contents are centrifuged to remove enzymes. Take the supernatant to count the cells, adjust the cell density according to the results of cell technology, and then perform single-cell sequencing. Single Cell Sequencing 10xGenomics Single Cell A Chip Kit, Single Cell 3'Library and Gel Bead KitV2 were used to perform 3' single cell RNA sequencing on the cell suspension of each sample, and the number of recovered target cells...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com