Novel CRISPR-Cas12i system
A wild-type protein technology, which is applied in the direction of urinary system diseases, muscular system diseases, nervous system diseases, etc., can solve the problems of good or bad performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0056] 1. A Cas12i protein, which comprises the amino acid sequence shown in any one of SEQ ID NO:1-10 (preferably, SEQ ID NO:1-3 and 6, more preferably, SEQ ID NO:1) Have at least 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%, Amino acid sequences that are 96%, 97%, 98%, 99%, 99.5% or 100% identical.
[0057] The Cas12i protein may also contain amino acid mutations that have no substantial effect on the catalytic activity (endonuclease cleavage activity) or nucleic acid binding function of Cas12i.
[0058] 2. The Cas12i protein according to any one of the preceding embodiments, wherein the Cas12i protein substantially lacks (for example, retains less than 50%, 40%, 35%, 30%, 27.5%, 25%, 22.5%, 20% %, 17.5%, 15%, 12.5%, 10%, 7.5%, 5%, 4%, 3%, 2.5%, 2%, 1% or lower) corresponding to wild-type Cas12i protein targeting and guide sequence Spacer-specific endonuclease cleavage activity of the target sequence of complementary target DNA.
[0059] ...
Embodiment 1
[0272] Embodiment 1: the identification of novel Cas12i protein
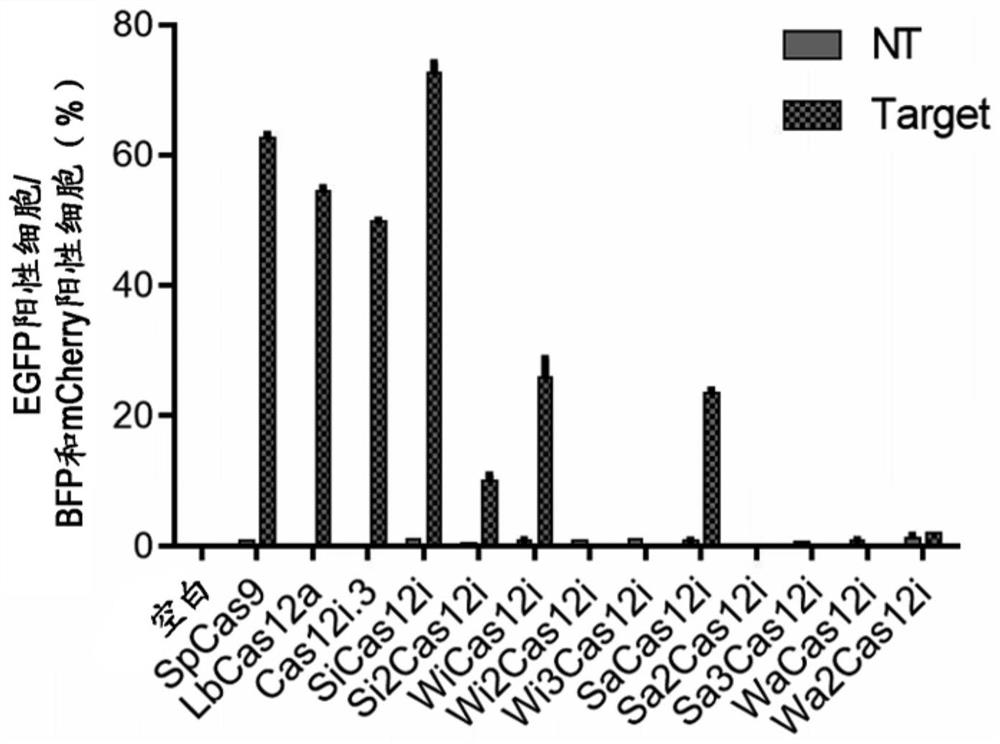
[0273] Download the metagenomic database (https: / / jgi.doe.gov / ) of the Joint Genome Institute (JGI), and get a total of 16TB of high-quality data. Over 6,000 metagenomic datasets were locally aligned with TBLASTN (https: / / blast.ncbi.nlm.nih.gov / ). Among them, 10 new Cas12i proteins were found in 4 groups of samples with different origins, which were respectively named SiCas12i, Si2Cas12i, WiCas12i, Wi2Cas12i, Wi3Cas12i, SaCas12i, Sa2Cas12i, Sa3Cas12i, WaCas12i and Wa2Cas12i. SiCas12i is 287 and 193 amino acids smaller than SpCas9 and LbCas12a, respectively.
[0274] The amino acid sequences of SiCas12i, Si2Cas12i, WiCas12i, Wi2Cas12i, Wi3Cas12i, SaCas12i, Sa2Cas12i, Sa3Cas12i, WaCas12i and Wa2Cas12i are respectively SEQ ID NO: 1-10, and the nucleotide coding sequences are respectively SEQ ID NO: 11-20. Four Cas12i-containing samples were annotated with CRISPR loci using PILERCR, and the DR sequences correspondi...
Embodiment 2
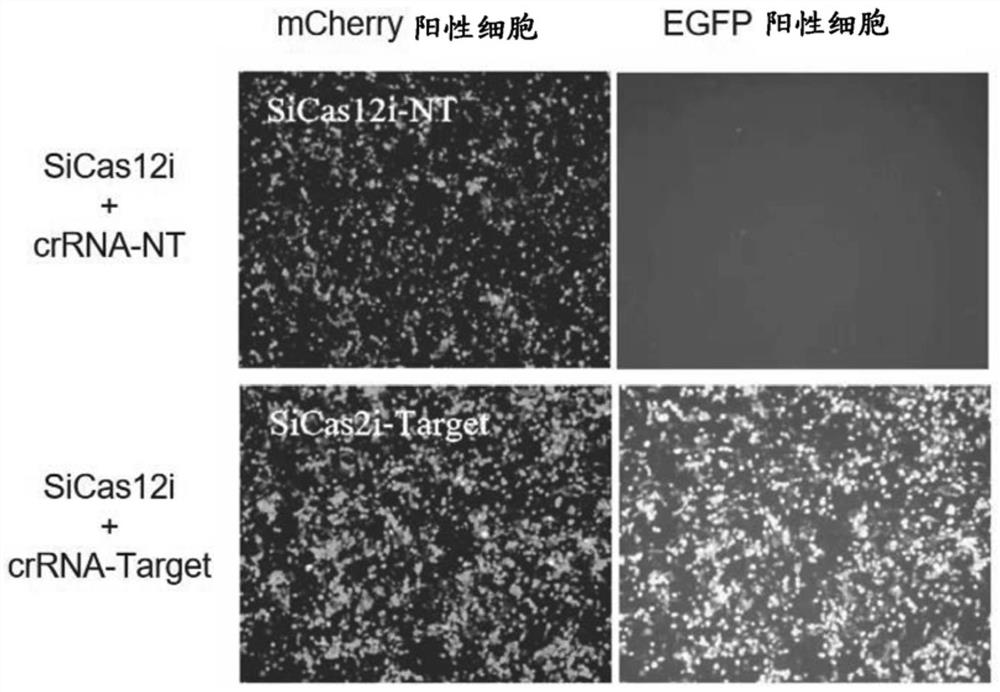
[0275] Example 2: Using a fluorescent reporter system to compare the cleavage activity of Cas12i and three kinds of control SpCas9, LbCas12a, Cas12i.3
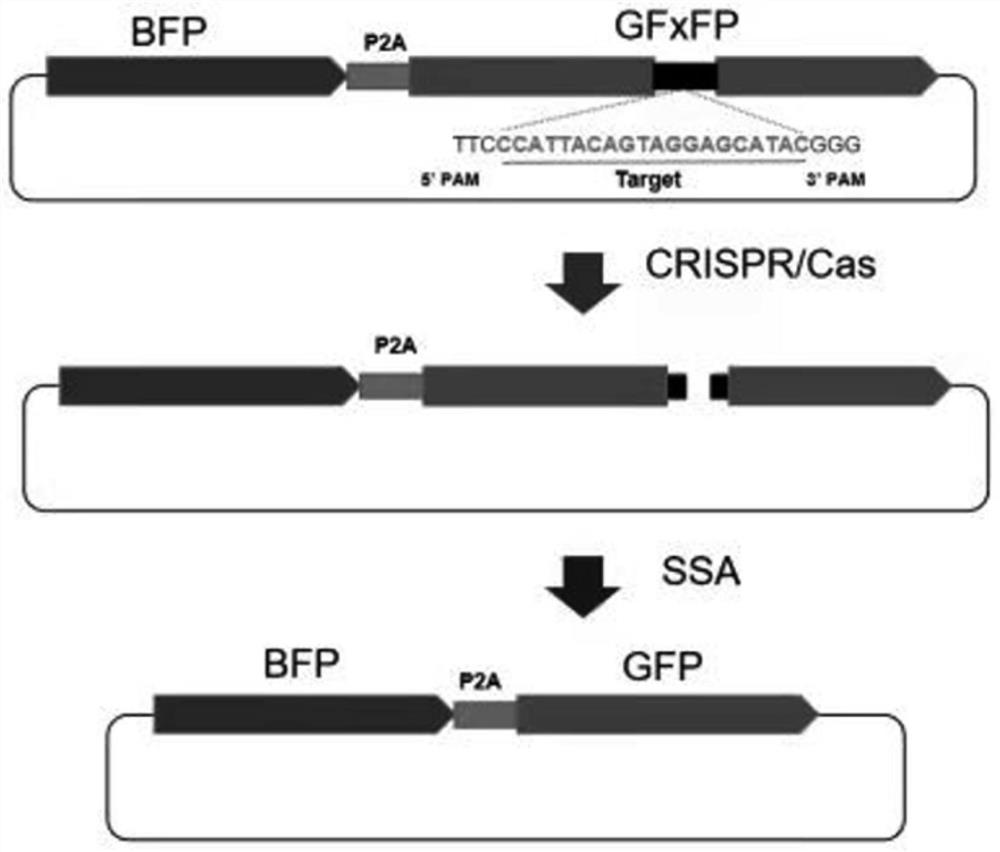
[0276] In order to sensitively detect the cleavage activity of the CRISPR / Cas system, a reporter vector for transcribing BFP-P2A-GFxFP mRNA (BFP-P2A-GFxFP reporter vector, SEQ ID NO:31, figure 1 ). BFP expresses the coding sequence of blue fluorescent protein, and the blue fluorescence indicates that the reporter vector has successfully transfected the host cell. The GF and FP sequences in GFxFP are the N-terminal 561nt sequence and the C-terminal 609nt sequence of the green fluorescent protein gene EGFP, respectively, and the two sequences have a total overlap of 450nt. The middle of GFxFP is an insert fragment (SEQ ID NO:32), which contains the target sequence targeted by the CRISPR / Cas system (SEQ ID NO:33).
[0277] Most of the currently known Cas12 proteins recognize the 5'-T enriched PAM of double-stranded DNA, while C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com