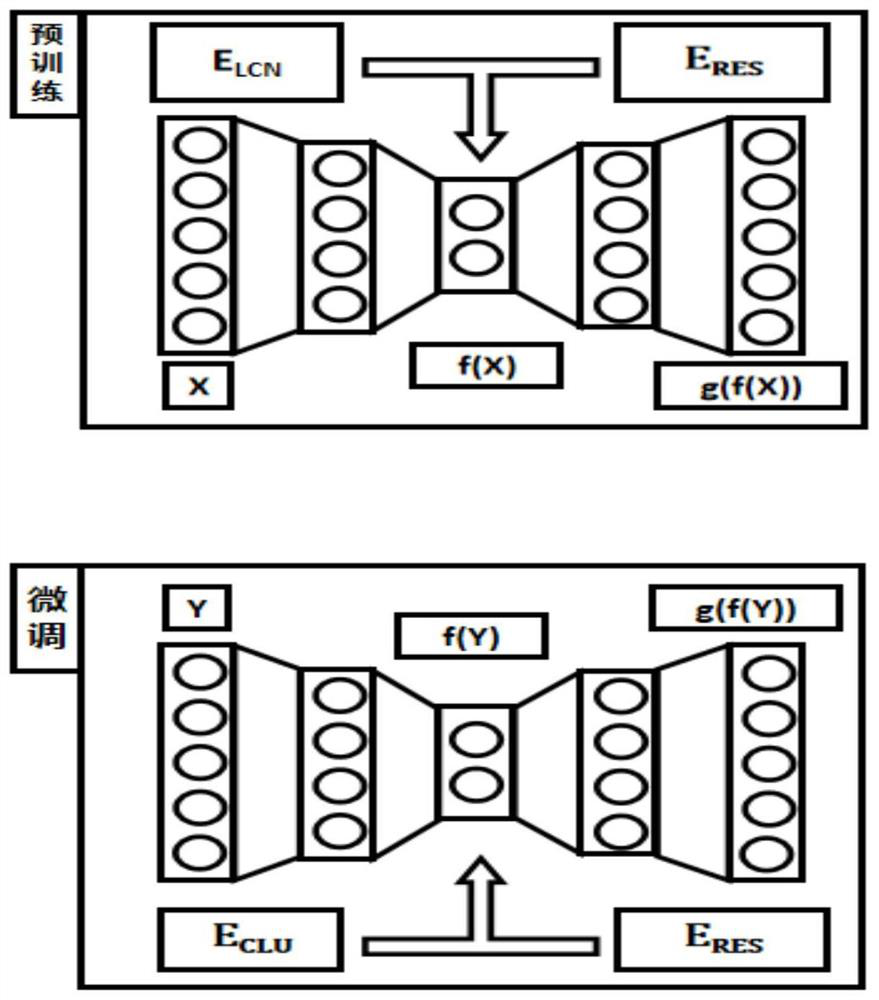
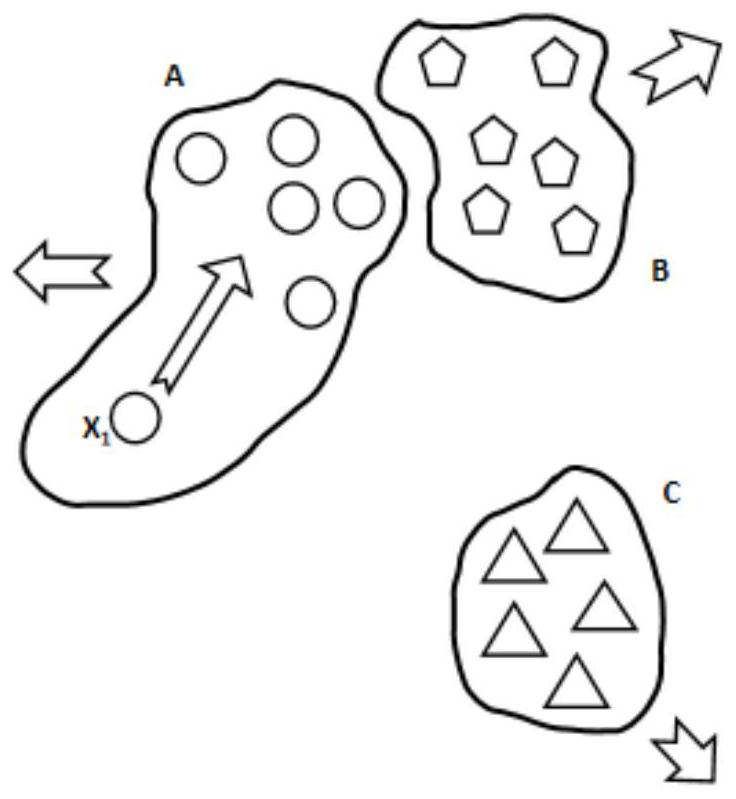
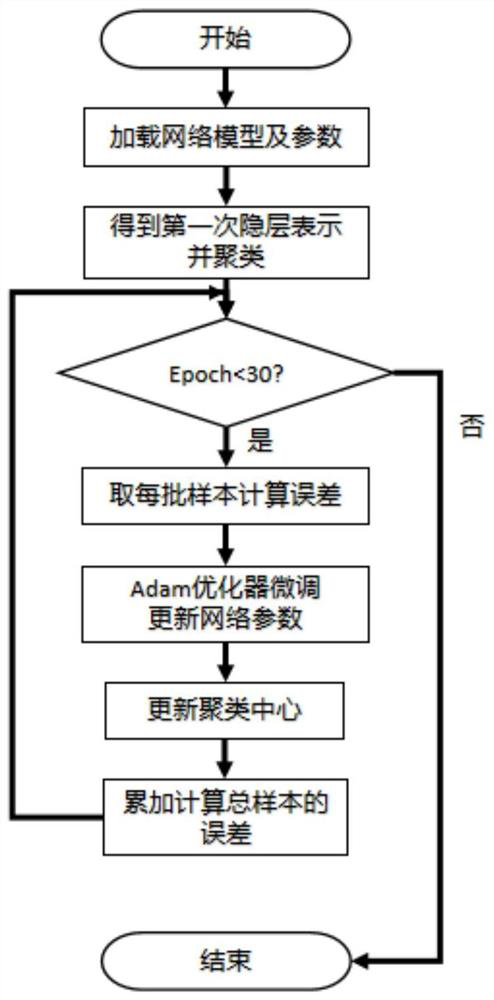
Metagenome sequence deep clustering method based on reference species tag constraint
A technology of metagenomic and clustering methods, applied in the field of deep clustering of metagenomic sequences based on reference species label constraints, can solve the problems of high similarity of adjacent species and inaccurate clustering, and achieve the effect of excellent clustering performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0178] In this embodiment, the test set includes two data sets, the Sharon simulation data set and the Strain simulation data set. The Sharon simulation data set contains 37628 contigs sequences of 101 species, which are simulated based on the first 96 data sets of the HMP project. The Strain simulation data set is the ability to test the taxonomic resolution of different species, containing 9401 contigs sequences of 20 species, including strains of the same species. The 20 species consisted of 5 different strains of Escherichia coli, 5 species of Bacteroides, 5 strains from different Clostridium species, and 5 strains of other typical intestinal bacteria.
[0179] Utilize the metagenome deep clustering method (Label-constrained deep clustering, LCDC) provided by the present invention to carry out analysis, and simultaneously use current similar analysis method COCACOLA, CONCOCT, MetaBAT and MaxBin2.0 as contrast, the analysis result is as follows Figure 5 , Figure 6 shown....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com