Application of nitrate transporter and coding gene thereof in increasing number of root nodules of hairy roots of crops
A technology for transgenic plants and hairy roots, which is applied in the field of genetic engineering to achieve the effects of improving the nitrogen-fixing capacity of nodules, increasing the number of nodules of hairy roots, and improving soybean yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Embodiment 1, gene
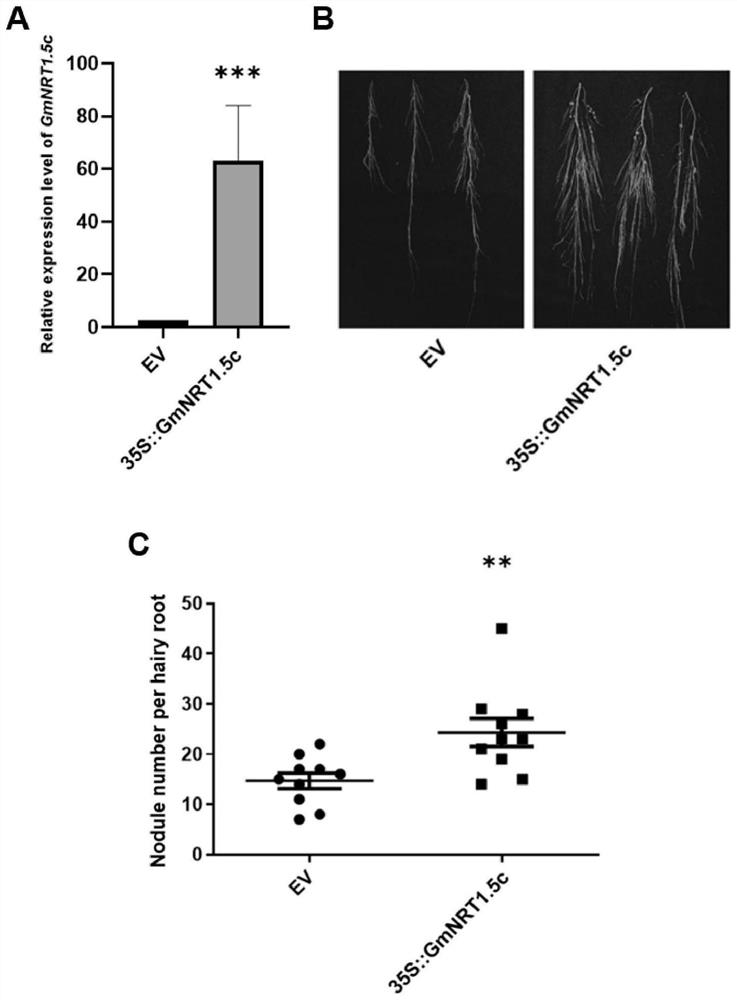
[0022] This example involves soybean GmNRT1.5c The gene, the nucleotide sequence of its cDNA reading frame is shown in SEQ ID NO:1. A sequence corresponding to this gene that is completely complementary to SEQ ID NO: 1, nucleotides that are substituted, deleted or increased by one or more nucleotides on the basis of the sequence shown in SEQ ID NO: 1, and have the same function sequence. Considering the degeneracy of codons, without changing the amino acid sequence, appropriately modifying the base sequence of the above cDNA reading frame is also a manifestation of the technical solution of the present invention.
Embodiment 2
[0023] Embodiment 2, protein
[0024] This example relates to the protein corresponding to the nucleotide sequence of Example 1. without affecting soybean GmNRT1.5c On the premise of protein structure and activity, soybean GmNRT1.5c Various substitutions, deletions or additions of one or more amino acids, or terminal modifications to the amino acid sequence of the protein are also a manifestation of the technical solution of the present invention.
Embodiment 3
[0025] Embodiment 3, the acquisition of gene sequence
[0026] soybean GmNRT1.5c The cDNA reading frame of the gene can be obtained by the following method: using the soybean variety Williams 82 in the laboratory as a material, extract its RNA, reverse transcribe it into cDNA, design primers based on the existing Williams 82 genome sequence to clone soybean GmNRT1.5c Gene CDS, the primers include the upstream primer shown in SEQ ID NO:2 and the downstream primer shown in SEQ ID NO:3. Use high-fidelity DNA enzymes for PCR amplification. The PCR reaction system is: cDNA template: 1 μL; front primer (F): 0.5 μL; back primer (R): 0.5 μL; buffer: 15 μL; ddH 2 O: 19 μL; dNTP: 5 μL Total reaction system 30 μL. Reaction conditions: 94 °C: 2 min, 98 °C: 20 s, 56 °C: 30 s, 68 °C: 2 min, 68 °C: 10 min, 16 °C: ∞; where, 98 °C: 20 s, 56 °C: 30 s, 68 °C: 2 min, 38 cycles in three steps. 30 μL of PCR amplification products were analyzed by 1% agarose gel electrophoresis. The band of a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com