Gastric cancer metabolism gene prognosis prediction method and device
A prediction method and prediction device technology, which is applied in character and pattern recognition, instruments, calculations, etc., can solve the problems of large number of genes and lack of further exploration of the metabolic characteristics of gastric cancer, and achieve strong robustness and stable prediction performance , the effect of good prognosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
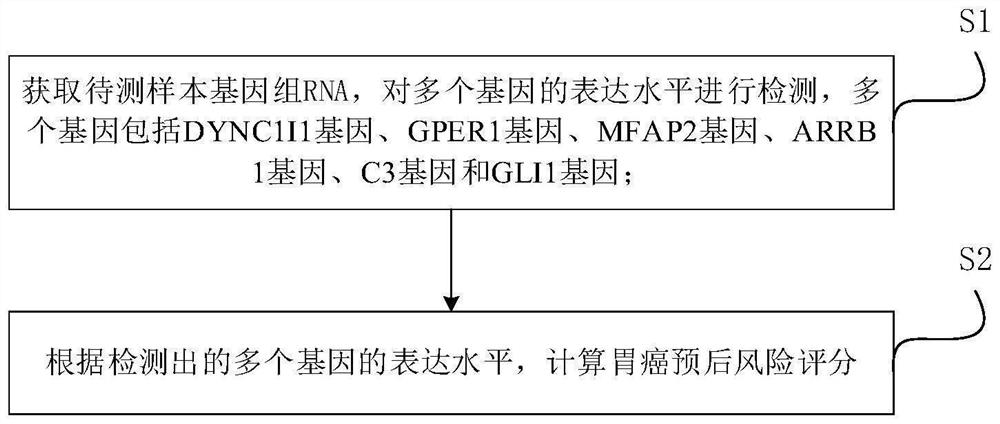
[0042] Such as figure 1 As shown, this embodiment provides a method for predicting the prognosis of gastric cancer metabolic genes, comprising the following steps:
[0043] S1: Obtain the sample to be tested, and detect the RNA expression levels of multiple genes, including DYNC1I1 gene, GPER1 gene, MFAP2 gene, ARRB1 gene, C3 gene and GLI1 gene;
[0044] S2: Calculate the gastric cancer prognosis risk score according to the detected expression levels of multiple genes.
[0045] Most preferably, the calculation expression of the gastric cancer prognosis risk score is:
[0046] RiskScore 6 =0.38585*exp DYNC1I1 +0.10411*exp GPER1 +0.04476*exp MFAP2 -0.70386*exp ARRB1 +0.09187*exp C3 +0.21797*exp GLI1
[0047] In the formula, RiskScore 6 is the prognostic risk score for gastric cancer, exp DYNC1I1 is the expression level result of DYNC1I1 gene based on the natural constant e, exp GPER1 is the expression level result of GPER1 gene based on the natural constant e, exp M...
Embodiment approach
[0048] As an optional implementation, the method further includes: loading the detected expression levels of multiple genes into a pre-established and trained classifier, calculating the gastric cancer prognostic risk score, and calculating the gastric cancer prognostic risk score greater than the preset The samples to be tested at the risk threshold are divided into a high-risk group, otherwise they are divided into a low-risk group.
[0049] As a preferred embodiment, the method also includes:
[0050] In this example, calculate the RiskScore 6 After that, the RiskScore 6 Perform Z-score transformation into Risk Score (data standardization), and divide samples with Risk Score greater than zero into the high-risk group, and samples less than zero into the low-risk group.
[0051] This embodiment also provides a gastric cancer metabolic gene prognosis prediction device, including:
[0052] The data acquisition module is configured to: acquire the expression levels of multip...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com