Third-generation sequencing RNA-seq comparison method based on WFA algorithm
An algorithm and sequencing technology, applied in the field of sequence comparison in bioinformatics, to achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
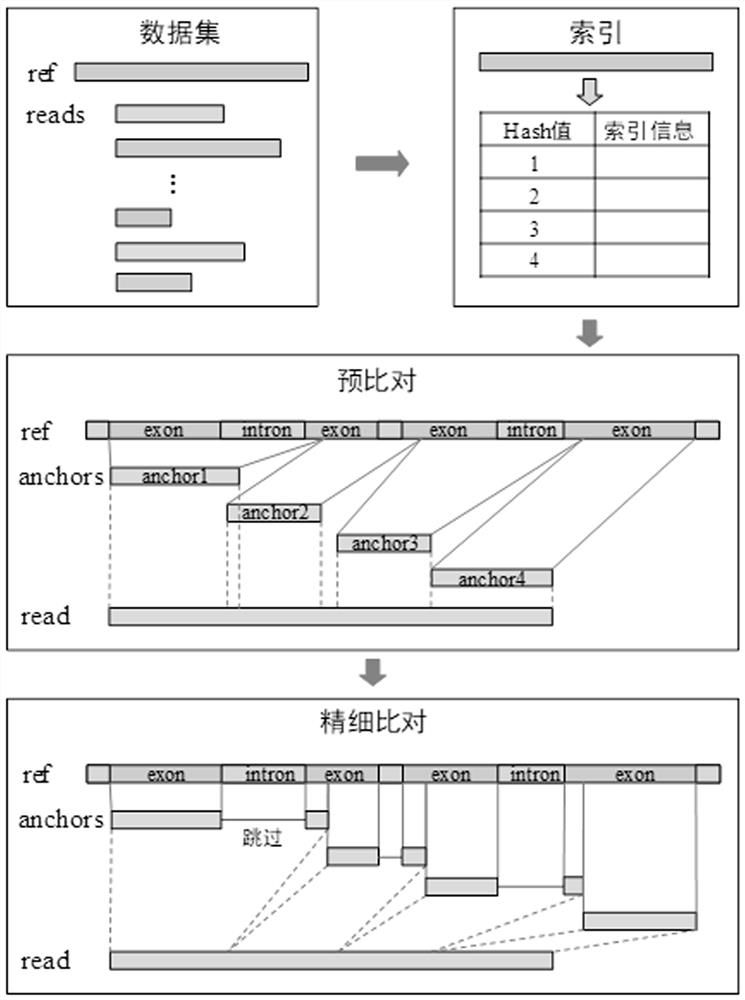
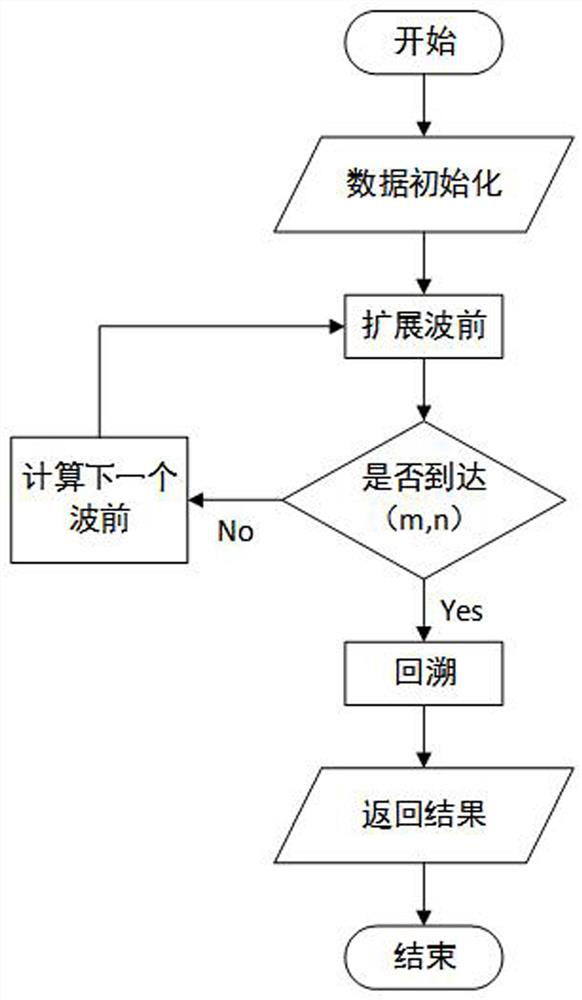
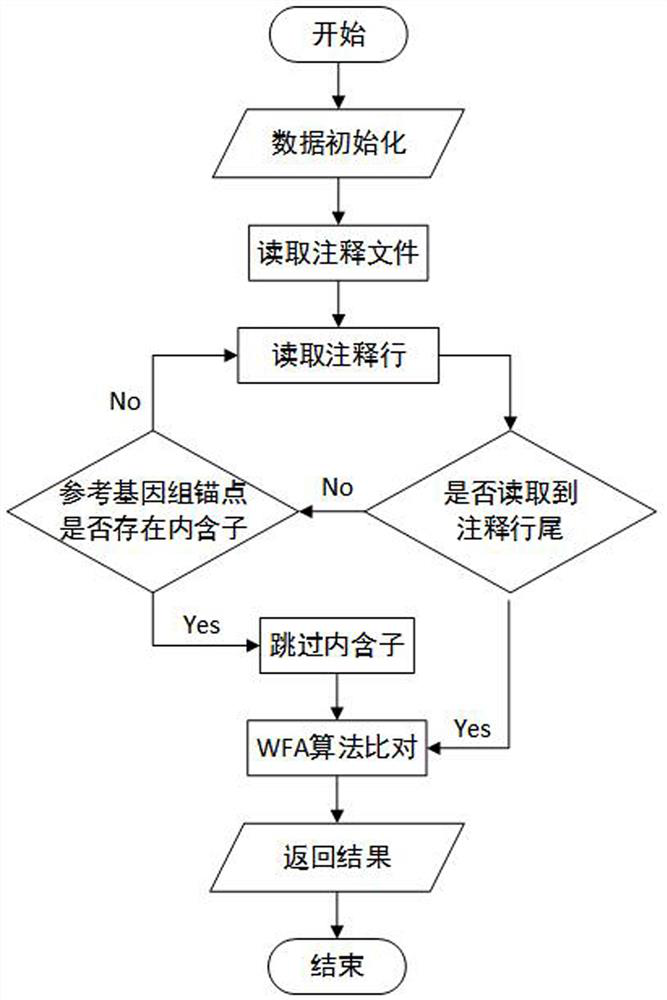
[0037] A third-generation sequencing RNA-seq alignment method based on the WFA algorithm, comprising the following steps:
[0038] (1) Obtain the data sets required for the evaluation, including 4 simulated data sets and 3 real data sets. The 4 simulated data sets were simulated and generated using the PBSIM tool, which simulated PacBio ROI Saccharomyces cerevisiae, Drosophila melanogaster, and human No. 19 respectively. Chromosome and ONT R2 2D Drosophila melanogaster data; 3 real datasets include RacBio ROI Drosophila melanogaster, error-corrected PacBioROI Drosophila melanogaster, and PacBio subreads of Drosophila melanogaster. Here, data from different species and different technologies are used to evaluate the performance of the algorithm more comprehensively. Table 1 shows the information of third-generation sequencing RNA-seq reads. For different species, each species corresponds to a reference genome ref. Use ref and reads to compare to find the position of reads in ref;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com