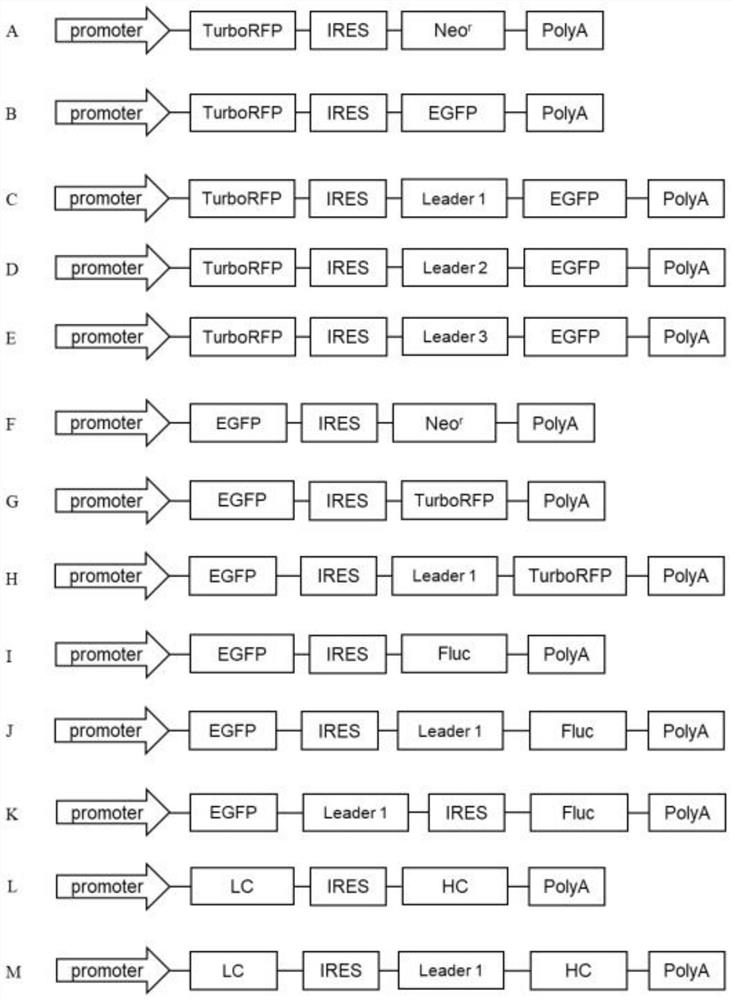
Leader sequence for improving non-cap-dependent translation efficiency and application thereof
A leading sequence and high-efficiency technology, applied in the field of genetic engineering, can solve problems such as low efficiency of downstream gene expression, protein heterogeneity, and incomplete cutting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] 1. Construction of monocistronic vector containing TurboRFP gene
[0028] (1) Synthesis of TurboRFP gene
[0029] Referring to the TurboRFP gene sequence (GenBank: MW560964, bases 5851 to 6546), the TurboRFP gene was synthesized, which was specifically handed over to General Biological Gene (Anhui) Co., Ltd. for synthesis. The synthesized TurboRFP gene is completely consistent with the TurboRFP sequence published by GenBank (GenBank: MW560964, bases 5851-6546). In order to facilitate cloning, when synthesizing the TurboRFP gene sequence, a 5'-AGCAAGCTT-3' sequence was introduced at the 5' end, where AGC was a protective base, AAGCTT was a HindIII restriction site, and 5'-ATAGCGGCCGC-3' was introduced at the 3' end. , wherein ATA is the protective base, and GCGGCCGC is the Not I restriction site.
[0030] (2) Construction of expression vector containing TurboRFP sequence
[0031] The PCR amplification product of TurboRFP double digested with HindIII and Not I, and the...
Embodiment 2
[0065] 1. Construction of a monocistronic vector containing EGFP
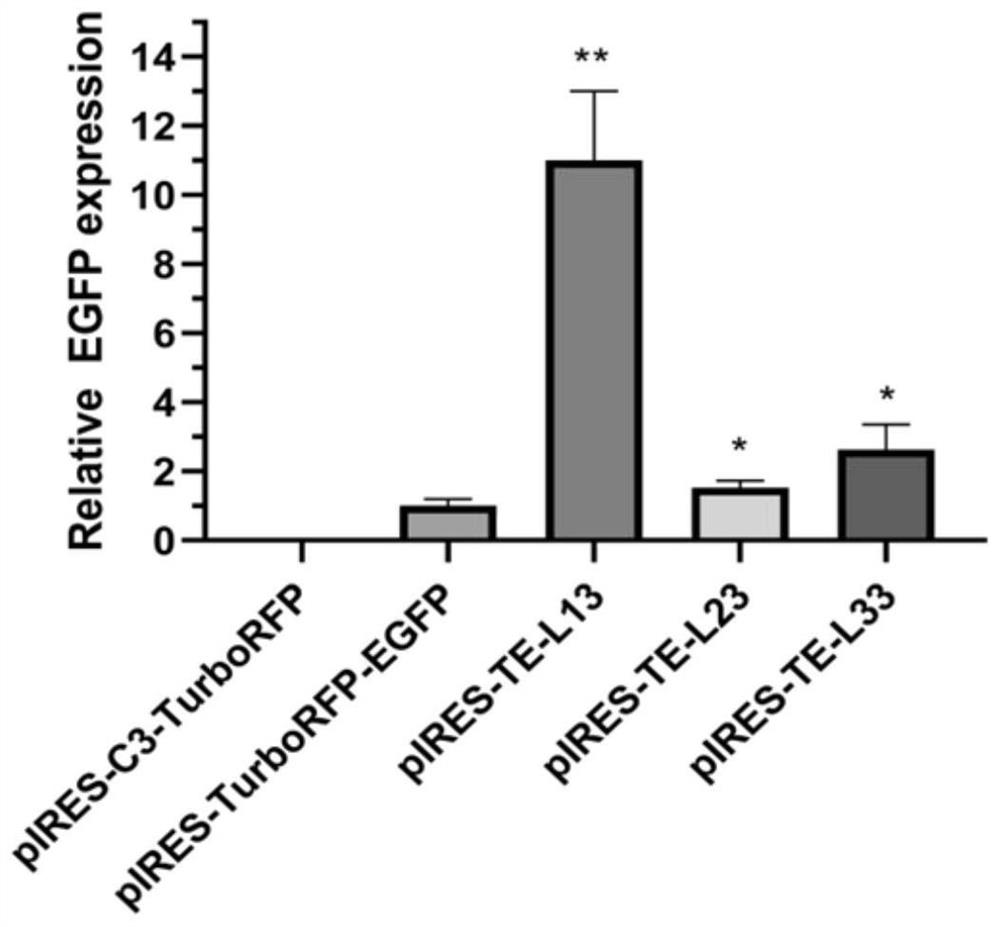
[0066] Amplify EGFP according to the method of Example 1, and the method for constructing the EGFP monocistronic vector is the same as the method for constructing the TurboRFP gene monocistronic vector, except that the EGFP gene is used to replace the TurboRFP gene to obtain the vector pIRES-C3-EGFP, which The structure diagram is as figure 1 shown in F.
[0067] 2. Construction of pIRES-EGFP-TurboRFP gene bicistronic vector
[0068] (1) PCR amplification of TurboRFP gene
[0069] The primers P3 and P4 were designed with reference to the pIRES-C3-TurboRFP sequence constructed in Example 1, and SmaI and XbaI restriction sites were introduced into the 5' ends of the primers, respectively. The primer sequences are as follows (underlined are restriction sites):
[0070] P3: 5'-CCGCCCGGGATGAGCGAGCTGATCAAGGAGAAC-3' (as shown in SEQ ID NO. 6);
[0071] P4: 5'-CTATCTAGATCATCTGTGCCCCAGTTTGCTA-3' (as shown in SEQ ID ...
Embodiment 3
[0086] In order to verify whether the leader sequence can improve the expression of Fluc and in order to increase the fold accurately and accurately, the pIRES-EGFP-Fluc bicistronic vector and pIRES-EF-L13 containing the leader sequence (SEQ ID NO. 1) were constructed in this example. vector.
[0087] 1. Construction of pIRES-EGFP-Fluc bicistronic vector
[0088] The Fluc gene was artificially synthesized with reference to the gene sequence of Firefly luciferase (Fluc) (GenBank: MK484108.1, bases 648-2300), which was specifically handed over to General Biogene (Anhui) Co., Ltd. for synthesis. The synthesized Fluc gene is consistent with GenBank: MK484108.1, the 648-2300th nucleotide sequence.
[0089] The pIRES-EGFP-Fluc bicistronic vector was constructed with reference to the method for constructing the pIRES-EGFP-TurboRFP vector in Example 2, and the difference from Example 2 was that the TurboRFP gene in Example 2 was replaced with the Fluc gene. The schematic diagram of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com