Oligouncleotide for detection and identification of mycobacteria
A technology of oligonucleotide and mycobacteria, applied in the field of nucleotide sequence of ITS, can solve the problem of no distinction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0054] Below will introduce some better examples of the present invention and its reference accompanying drawing through embodiment. Example 1. Mycobacterial Culture and Genomic DNA Isolation
[0055] Mycobacterium standard strains come from KCTC (Korea Type Culture Collection) and ATCC (American Type Culture Collection), and clinical strains come from the Korean National Tuberculosis Association, National Masan Tuberculosis Hospital and Pusan National University Hospital clinical microbiology experts Manage the storage and cultivation of strains. The mycobacteria and pathogens used in the examples are listed in Table 8.
example 2
[0056] The extraction process of mycobacterial DNA was as follows: Mycobacteria were cultured in Ogawa medium. Then, the strain cultured with a platinum ring was placed in a microcentrifuge tube, mixed with 200 μl of InstaGene matrix (Bio-Rad Co.), and incubated at 56° C. for 30 minutes. After an additional 10 minutes of vortex mixing, the mixture was placed at 100°C for 8 minutes. After an additional 10 minutes of vortex mixing, the mixture was centrifuged at 12,000 rpm for 3 minutes. Take the supernatant and put it in another test tube, and store it at -20°C. In the following PCR, 2 μl of the DNA solution of each strain will be used. Example 2. Amplification of Mycobacterial ITS Primers for Manufacturing
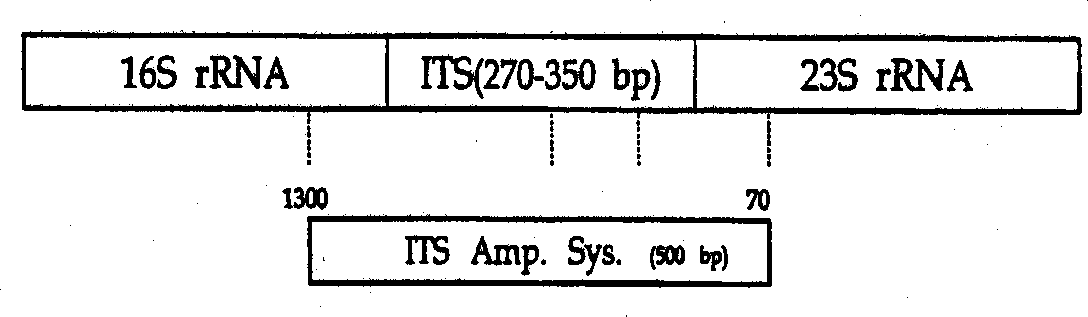
example 3
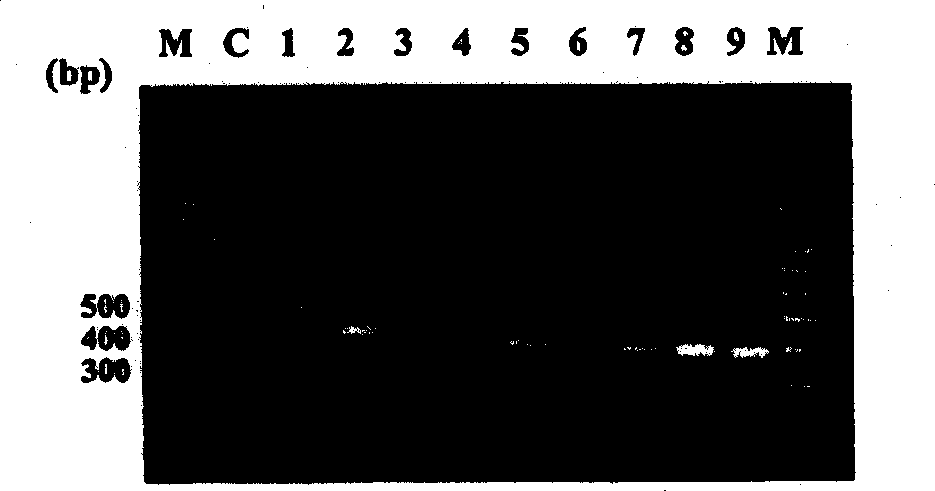
[0057] Mycobacterial ITS and the primers used to amplify ITS by PCR are listed in figure 1 middle. Primers for amplifying mycobacterial ITS were constructed based on several fragments of mycobacterial 16SrRNA and 23SrRNA conserved regions. The DNA sequences of 16SrRNA and 23SrRNA of mycobacteria in the gene bank were analyzed by multiple sequence alignment and immature cell inspection. Then, primers were designed through a part of DNA of 16SrRNA and 23SrRNA, thereby selectively amplifying the ITS region of about 500 bp. The DNA sequences of these primers are listed in SEQ ID NOs: 242 (IT SF) and 243 (ITSR). All primers used in this example were prepared by Derkin-Elmer DNA synthesizer (BioBasic, Canada) at a concentration of 50 nmol. Example 3. PCR and product identification
[0058] 2 µl of the DNA solution in Example 1 was used in PCR. The reaction solution includes: 500mM KCl, 100mM Tris HCl (pH9.0), 1% Triton X-100, 0.2mM dNTP (dATP, dGTP, dTTP and dCTP), 1.5mM MgCl 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com