Circadian control of stem/progenitor cell self-renewal and differentiation and of clock controlled gene expression
A technology of clock control and stem cells, applied in the field of rhythm control system
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 and 2
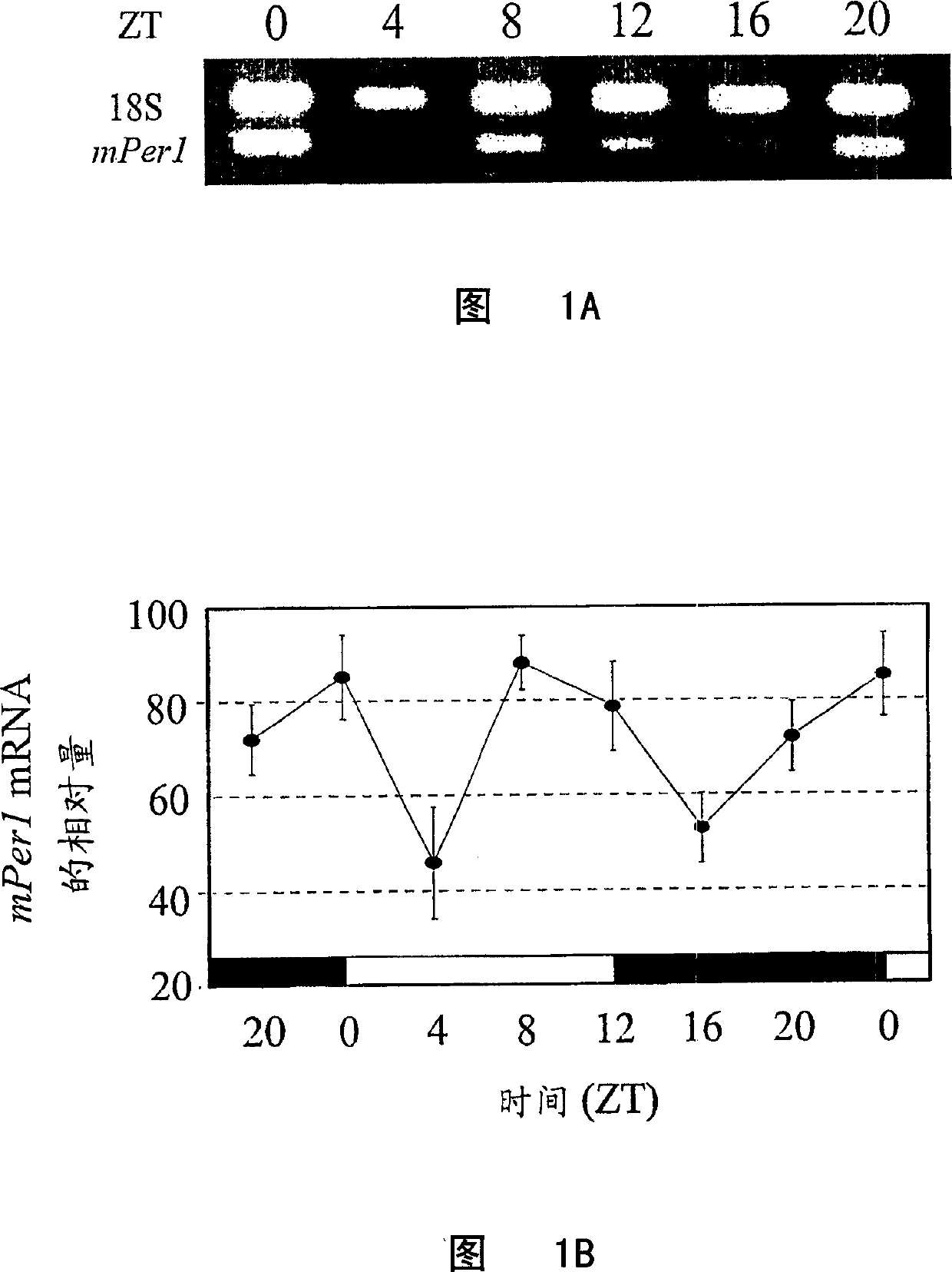
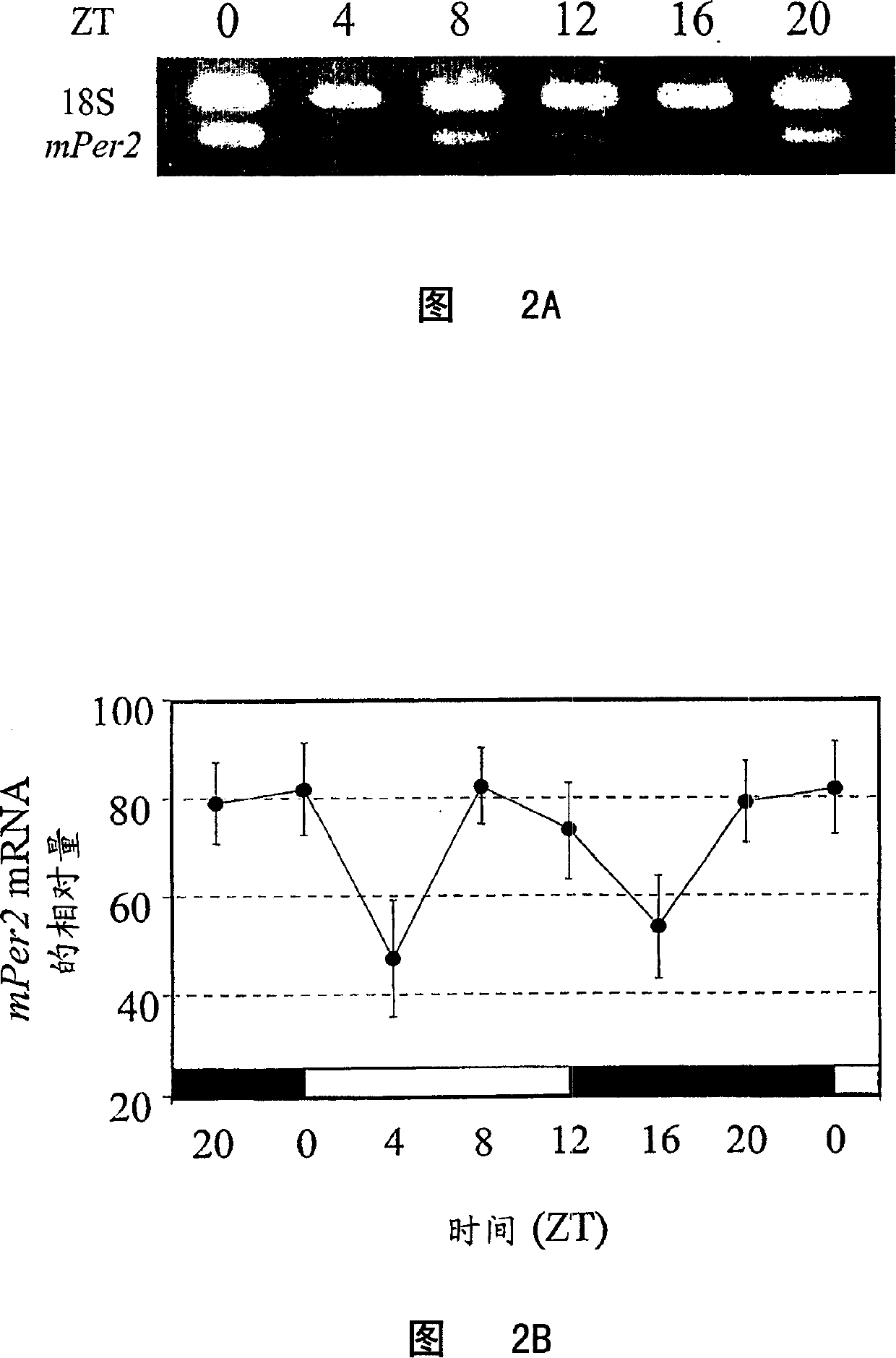
[0099] In RT-PCR experiments, samples collected at six time points were analyzed each time. Total RNA was purified from Gr-1 positive or non-isolated bone marrow cells using RNeasy MiniKit (Qiagen) according to the instructions. After DNase (Promega) treatment, approximately 2 μg of total RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (MMLV-RT; Stratagene). The reverse transcriptase was then inactivated at 90°C for 5 minutes. According to the instructions, the relative quantitative analysis of mPer1 and mPer2 mRNA at different time points was performed using an internal control (quantitative RNA18S internal standard; Ambion). The 18S non-replication-competing primer (Competimer; Ambion) carries a modified 3' end that blocks DNA polymerase extension. The ratio of 18S non-replication competing primer to 18S primer was 9:1 to reduce the amount of 18S cDNA signal relative to the target gene. 18S cDNA and target cDNA (mPer1 or mPer2) were co...
Embodiment 3
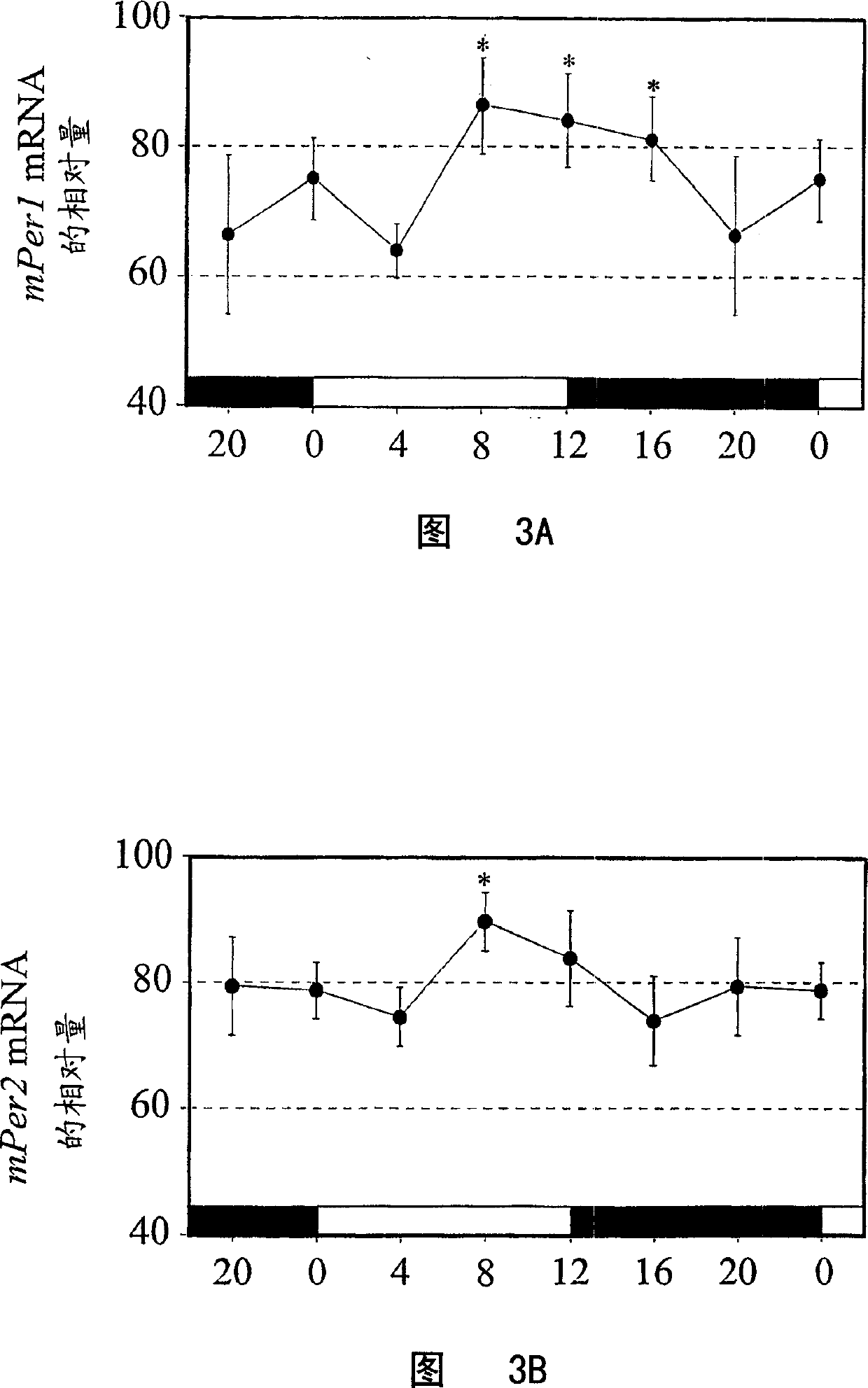
[0115] In RT-PCR experiments, samples collected at six time points were analyzed each time. Use RNeasy MiniKit (Qiagen), follow the instructions, from lin - Or purify total RNA from unisolated bone marrow cells. With SUPERSCRIPT II reverse transcriptase (Gibco), random primers ((Invitrogen, Carlsbad, CA) pairs from 10 6 unseparated bone marrow cells and 2×10 5 a lin - The total RNA extracted from the cells was reverse-transcribed in 20 μl, and the reaction condition was 42° C. for 60 minutes. According to the instructions, the relative quantitative analysis of mPer1, mClock or GATA-2 mRNA at different time points was performed using an internal control (quantitative RNA18S internal standard; Ambion). The 18S non-replication-competing primer (Competimer; Ambion) carries a modified 3' end that blocks DNA polymerase extension. The ratio of 18S non-replication competing primer to 18S primer was 10:1 to reduce the amount of 18S cDNA signal relative to the target gene. 18S cDN...
Embodiment 3 and 4
[0137] The luciferase reporter construct was performed as follows: clone 3a-7 was digested with KpnI and Sad and the excised insert was cloned into the same site of pGL3-Basic ((Promega, Madison, WI) to construct pGL3-3a-7 The DNA fragment between the EcoRI site and the third PmlI site or the first PmlI site and the XbaI site (5' to 3') of pGL3-3a-7 was removed to generate pGL3-3a-31, respectively Or pGL3-3a-39. The pGL3-Elb reporter vector is derived from pG5Elb-Luc (Hsiao et al., "Kennedy's neurogenic disease and the first identified coactivator coupled to the polyglutamine region of the androgen receptor The link between ARA24", J. Biol. Chems ., 274 (29): 20229-20234 (1999), fully cited here, but 5 of the GAL4 binding sites were replaced with the multiple cloning sites of the pBluescript II KS (-) vector (Stratagene) (from KpnI to XbaI). Nucleotides corresponding to 76-351 in Figure 4 were PCR amplified and cloned into the EcoRI and BamHI sites of pGL3-Elb to generate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com