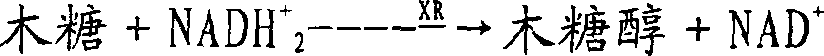Method for the biotechnological production of xylitol
A biotechnology, xylitol technology, applied in the field of biological preparation of xylitol, can solve the problem of not being able to effectively improve xylitol productivity, and achieve the effect of improving productivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] First, a preparation yeast strain expressing a heterologous xylose reductase is described.
[0049] Expression of heterologous xylose reductases in yeast is part of the prior art, described eg in WO91 / 15588.
[0050]Pichia stipitis was cultured overnight at 30° C. in 100 ml YPD (11 medium containing 10 g yeast extract, 20 g peptone and 20 g glucose, pH=6.5). Cells in 10 ml medium were centrifuged to pellet and isolate chromosomal DNA, according to the experimental design of Kaiser et al. (1994). This DNA was used as template for PCR amplification of the 956 bp intron-free open reading frame of the XYL1 gene (Amore et al. 1993). The resulting fragments were separated on a 1% agarose gel and purified using "JETQUICK Gel Extraction Spin Kit" from Genomed. The plasmids were treated with the corresponding restriction endonucleases and purified as described above. p425GPD was used as vector for expression in S. cerevisiae. This vector is a multicopy plasmid and contains t...
Embodiment 2
[0052] Inactivation of NADH dehydrogenase by gene replacement of NDE1 and NDE2
[0053] Cytosolic NADH dehydrogenase located in the mitochondria competes with XR for the cofactor NADH. To reduce cytosolic NADH consumption, the genes NDE1 and NDE2 were inactivated by gene replacement of appropriate DNA segments. The replacement fragment homologous to 40 bp at the 5' end of the NDE1 start codon and 40 bp at the 3' end of the stop codon was amplified by PCR. This fragment contains the his5+ gene of Schizosaccharomyces cerevisiae and complemented with the his3 mutation of S. cerevisiae (Wach et al. 1997). After amplification, this fragment was isolated and purified as described above. The replacement fragment was used to transform S. cerevisiae strain KOY50 according to the method of Schiestl and Gietz (1989). Isolation of histidine prototrophic transformants. Correct integration of this fragment and substitution with NDE1 was confirmed using diagnostic PCR. This strain (KOY5...
Embodiment 3
[0056] Gene replacement of the genes GPD1 and GPD2 encoding glycerol phosphate dehydrogenase
[0057] The cytoplasmic glycerol phosphate dehydrogenases GPD1p and GPD2p utilize NADH as a cofactor. Their inactivation preferentially results in the oxidation of cytosolic NADH by reducing xylose to xylitol using XR. At the time of gene replacement, Saccharomyces cerevisiae strains KOY50Δnde1 and / or KOY50Δnde1Δnde2 were used.
[0058] Both strains bear the ura3Δ0 mutation. For this cleavage, a fragment containing homology to GPD1 and GPD2 was used, as described in Example 2 for NDE1. A fragment containing the Candida albicans URA3 gene was used as a selectable marker. The DNA portions flanking this selectable marker are identical to each other (Goldstein et al. 1999). URA3 from Candida albicans complements ura3 mutations in Saccharomyces cerevisiae.
[0059] Cells with unimpaired uracil metabolism were sensitive to 5-fluoroorodic acid (5-FOA) in contrast to ura3 mutant cells. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com