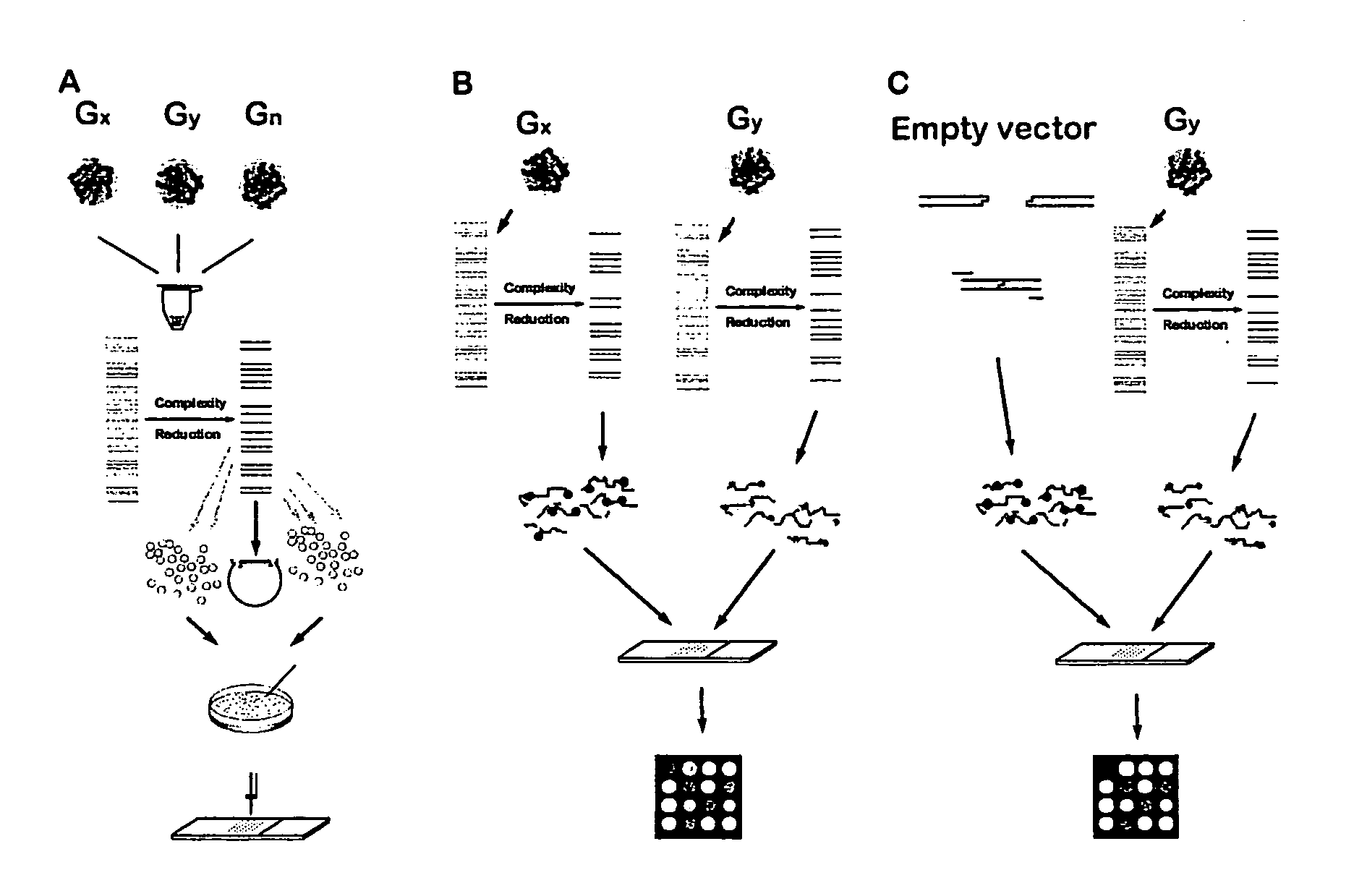
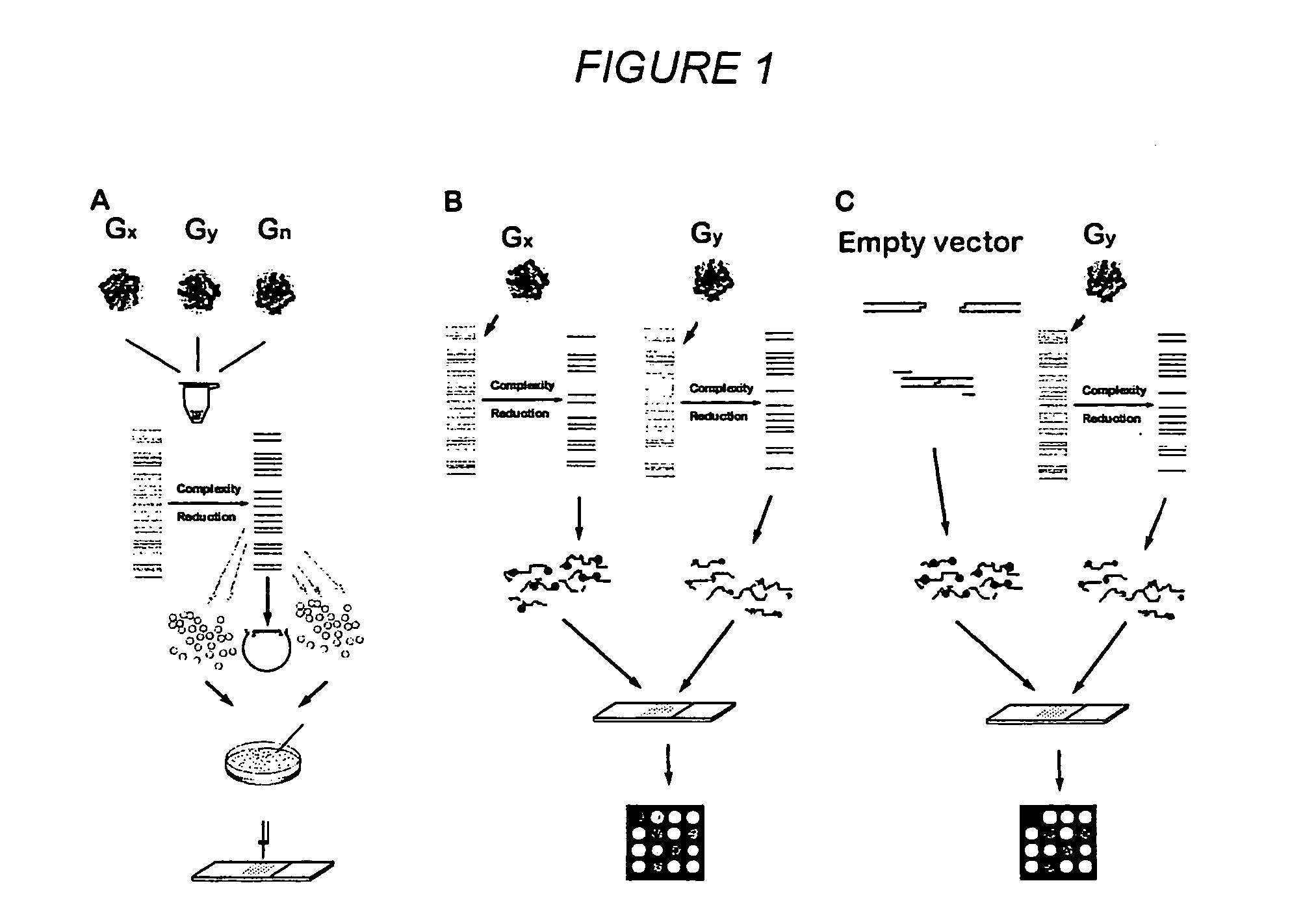
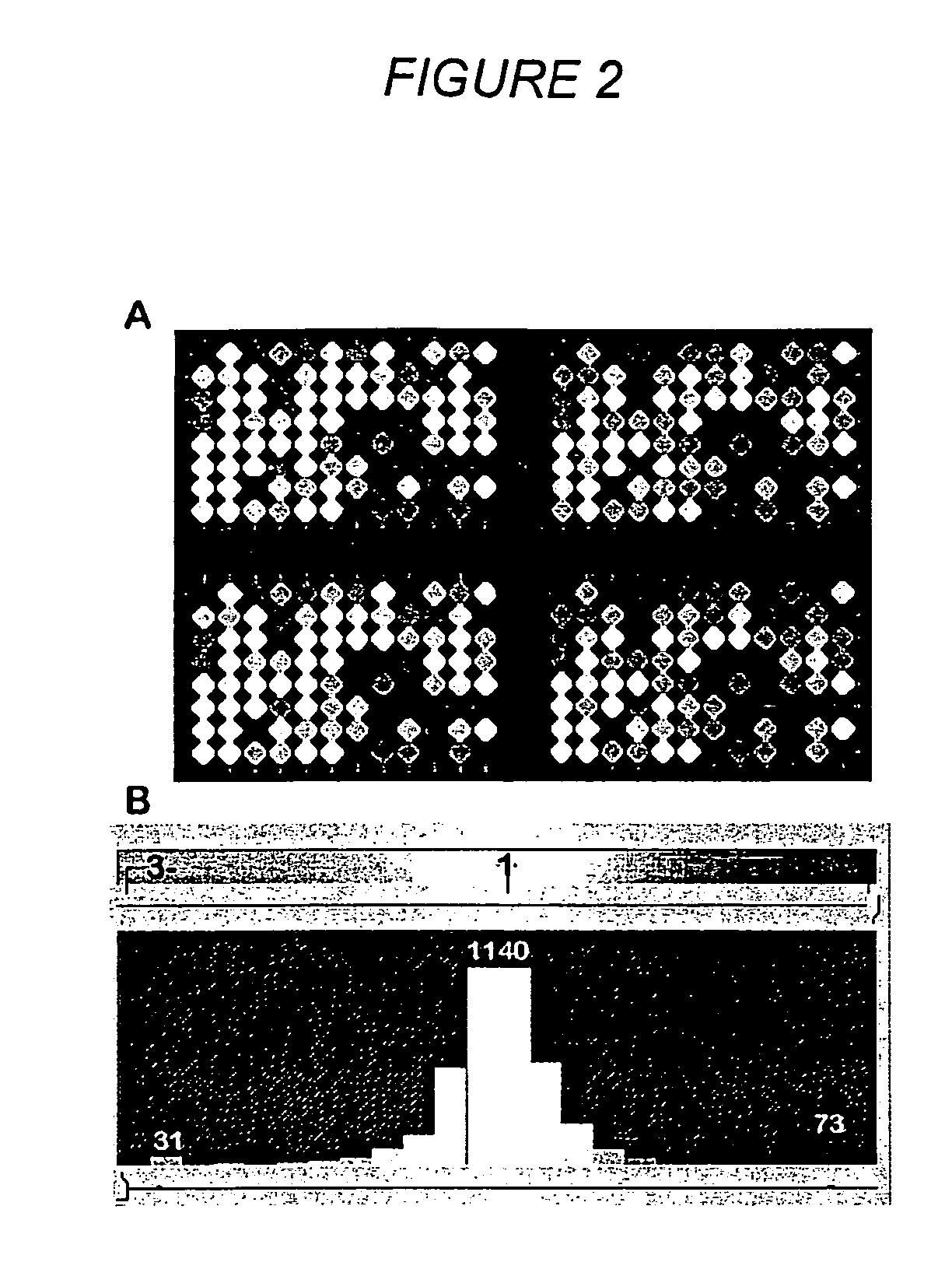
Methods for genotyping by hybridization analysis
a hybridization analysis and genotyping technology, applied in combinational chemistry, biochemistry apparatus, chemical libraries, etc., can solve the problems of additional difficulties in precisely correlating bands, inability to achieve, and laborious requirements of genotyping methods to analyze only a single organism, etc., to accelerate the speed of the introgression process and high throughput screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0114] Generating a Diversity Panel from Rice Genomic DNA
[0115] Representative samples of rice germplasm are identified for genotyping. The samples are chosen solely for demonstration purposes and are chosen on the basis of other knowledge for being a diverse set of genotypes. This is done mostly through analyzing dendrograms based on sequence and / or molecular marker polymorphism in order to pick up members of separate groupings. Also representative genotypes can be identified as representatives of separate clusters if the results (like Principal Component Analysis) or clustering algorithms are available. Alternatively representative genotypes can be identified through single pass sequencing of rapidly evolving segment of the genome followed by similarity / dissimilarity analysis. DNAs from a sampling of genotypes representing genetic diversity of rice species (usually 10-15) are used to generate DNA diversity panels through a number of techniques, one of which is exemplified below. ...
example 2
[0122] Preparation of a Diversity Array
[0123] The amplified DNA inserts are transferred into 384-well plates (Genetix) and arrayed using a microarrayer (e.g., 417 microarrayer; Affymetrix, Palo Alto, Calif.) onto Polysine™ microscope slides (MenzelGlazer, Germany) or in-house polylysine-coated microscope slides. Arrays are made with six replicates per fragment. The average center to center spot spacing is 250 μm.
[0124] At least 1 day after arraying, slides are processed by hydration in 1×SSC, quick drying, blocking for 15 min in a solution of NaBrH4 / PBS (prepared by dissolving 1 g NaBrH4 in 300 ml PBS, pH 7.0) (see also http: / / www.microarrays.org / protocols. html, Protocol for Post Processing Microarrays; June 2000, except that the succinate anhydride pyrolidone is replaced with NaBrH4 in PBS as the blocking solution). Slides are then dipped in boiling water for 30 sec to denature the DNA and followed by a 10 sec dip in 100% EtOH. Slides are dried by centrifugation at 1000 rpm in a...
example 3
[0125] Determination of Fingerprints by Hybridization of a Labeled Diversity Panel to an Array
[0126] For hybridization to a microarray prepared as taught in Example 3, a diversity panel of one or more specific genotypes is generated and labeled with a fluorescent dye. In a single hybridization experiment, a number of genotypes can be compared, the number being equal to number of labels that can be unequivocally detected and resolved. For example, an Affymetrix 418 scanner is equipped with “green” and “red” lasers, allowing for simultaneous analysis of two different samples.
[0127] Genomic DNA (200 ng-2 μg / μl) is cut with EcORI and ligated to EcORI adapters (1.5 μl of 5 pmoles / μl) using and excess of T4 ligase (40 units) for 3 hours at room temperature. For this step, 200 ng of DNA is sufficient, but 1 μg of DNA provides sufficient material for a number of hybridizations. Following ligation, the mixture is purified on a Qiagen column.
[0128] An amplification reaction contains 2.5 un...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com