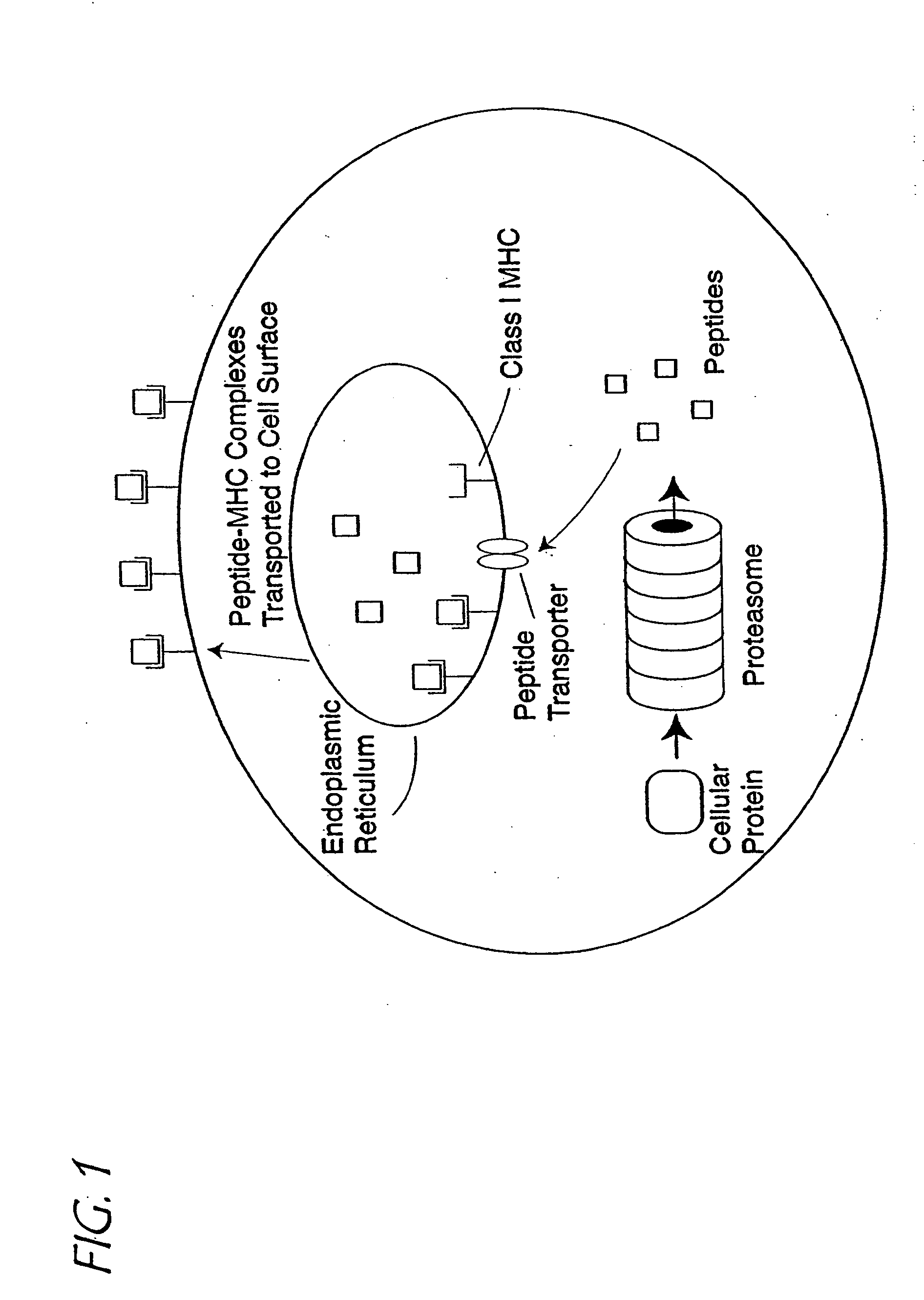
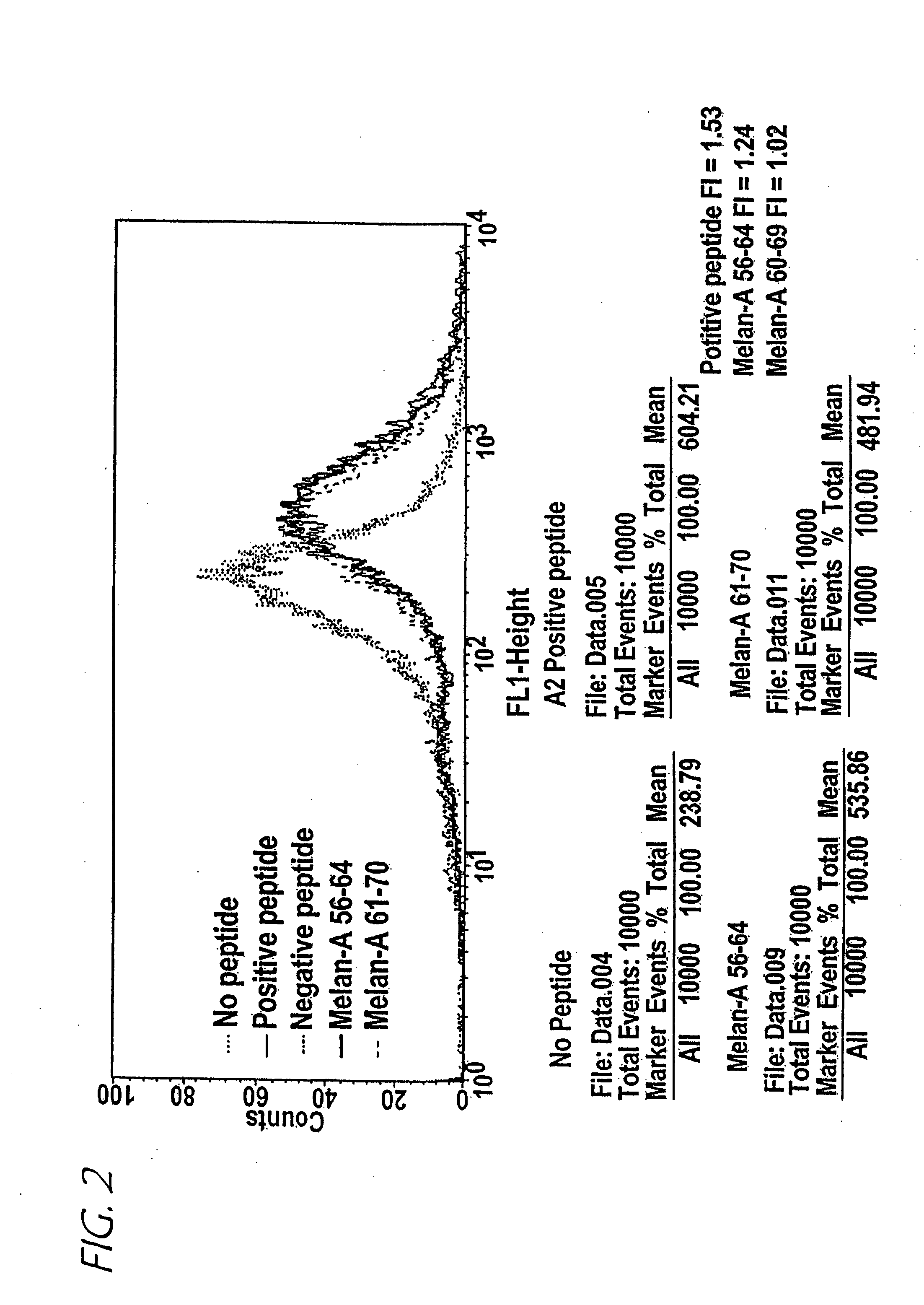
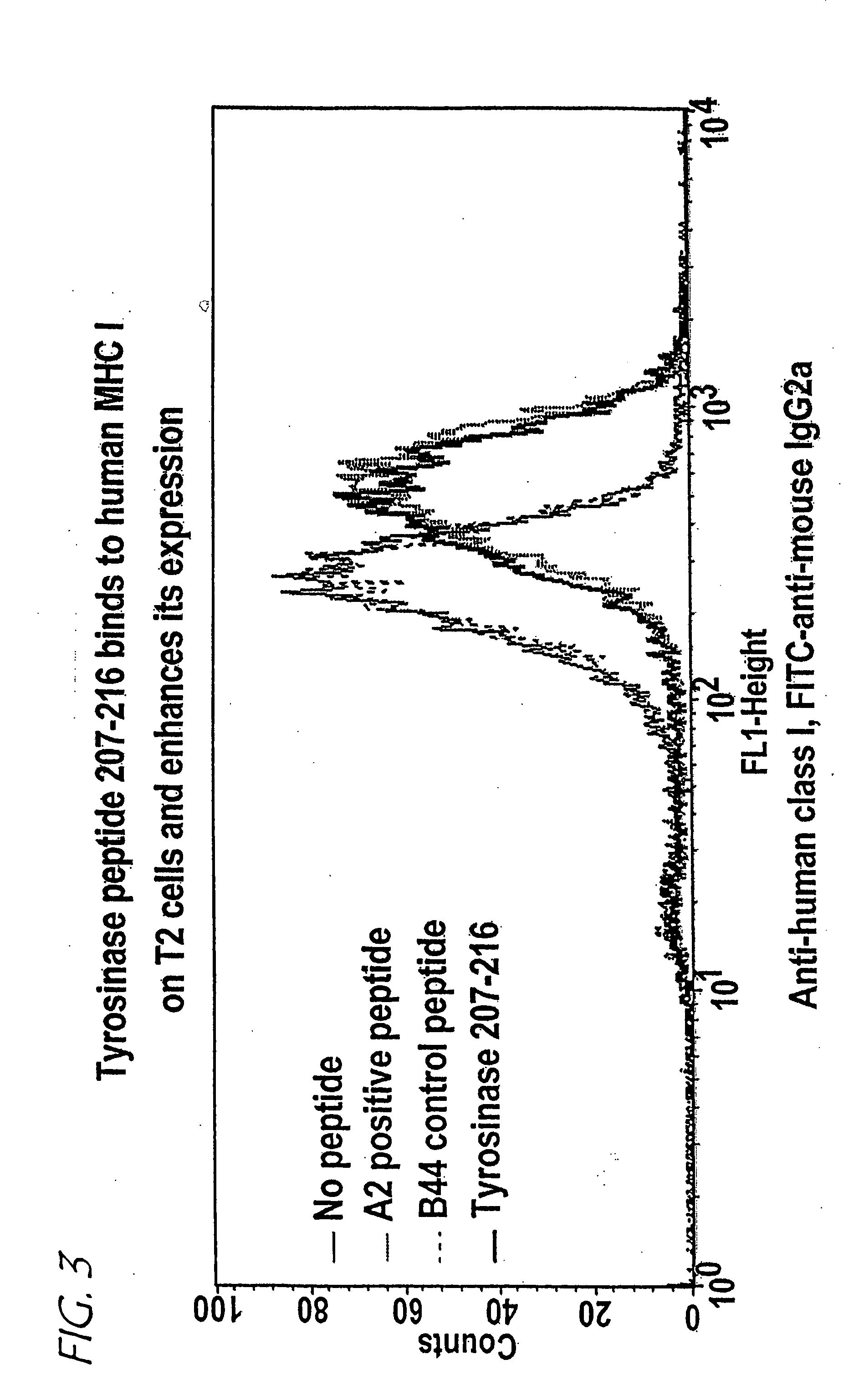
Method of epitope discovery
a technology of target cells and epitopes, applied in the field of target cell antigen identification, can solve the problems of host death, unable to effectively administer minimal epitopes for use as viral vaccines, and the ability of evading the immune system of the host,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Purification of Proteasome Complexes
[0102] A. Proteasome Complexes from Blood Cells
[0103] Concentrated erythrocyte bags were obtained from a local blood bank, (HemaCare, Van Nuys, Calif.). The contents of each bag were poured into 200 ml centrifuge tubes and washed 3 times with PBS by centrifugation at 2000 RPM for 10 minutes at room temperature in a swinging bucket rotor of a Megafuge 2.0 (Heraeus, Southplainfield, N.J.). After the last wash the samples were pooled in one container, to minimize variability among tubes, and then re-divided into several centrifuge tubes. The cells were centrifuged again at 2000 RPM for 10 min. The residual PBS was aspirated. The pellet was stored at −70° C. until use.
[0104] B. Proteasome Complexes from Tumor Cells
[0105] Raji cells, a Burkitt's lymphoma cell line, were obtained from ATCC, (American Type Culture Collection, Manassas, Va.). The cells were grown using standard cell culture methods and stimulated with INF-Gamma (100-500 U / ml) (Pharmin...
example 2
Generation of Predicted MHC I Peptide Cleft Binding Peptides Using Algorithmic Modeling
[0177] A population of candidate MHC I binding peptides, generated from the amino acid sequence of human carcinoembryonic antigen precursor (CEA) (GENBANK ACCESSION P06731), was produced using an algorithm. The particular algorithm is available at >, as discussed above and hereby incorporated by reference in its entirety. Once the algorithm was accessed, the amino acid sequence for CEA was provided. Next, parameters for the length of the epitope (decamers) and the particular MHC allele (H2-Db) of interest were selected. Following this, the data were submitted for algorithmic analysis. The resulting data are shown in Table II.
TABLE IIFragments of CEA having PredictedAffinity for H2-DbSeq IdPOS1234567890Scoreno+HZ,1 / 32547LQLSNGNRTL261369LQLSNDNRTL262191LQLSNGNRTL26353LLVHNLPQHL264371LSNDNRTLTL255549LSNGNRTLTL246193LSNGNRTLTL247299CQAHNSDTGL238100IIYPNASLLI219578SANRSDPVTL1910576SVSANRSDPV1911504S...
example 3
Digestion of Peptide Precursors Using Immune and Housekeeping Proteasomes to Determine Fragments Produced by Proteolytic Digestion
[0179] Peptides were synthesized using a 433A ABI synthesizer. Peptides were produced in 0.25 mmole quantities using Fastmoc chemistry. The peptides were tested for solubility and once solubilized, a 2 mM solution was prepared and divided into ˜25-30 μL aliquots which were stored at −20° C. for future use. Timed digest reactions, typically consisting of 2 μl of peptide and 4 μl of proteasome, were conducted with t=0 as a control and an incubation of the peptide with water instead of the proteasome as a further control. The reaction was carried out at 37° C. and ended by the addition of 10% TFA (trifluroacetic acid) on dry ice. The frozen samples were then analyzed by MALDI-TOF mass spectroscopy (MS) as described in Example 4, below.
[0180] An optional desalting step can be performed on the digests prior to MS analysis using the ZIP-TIP method (Millipore,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com