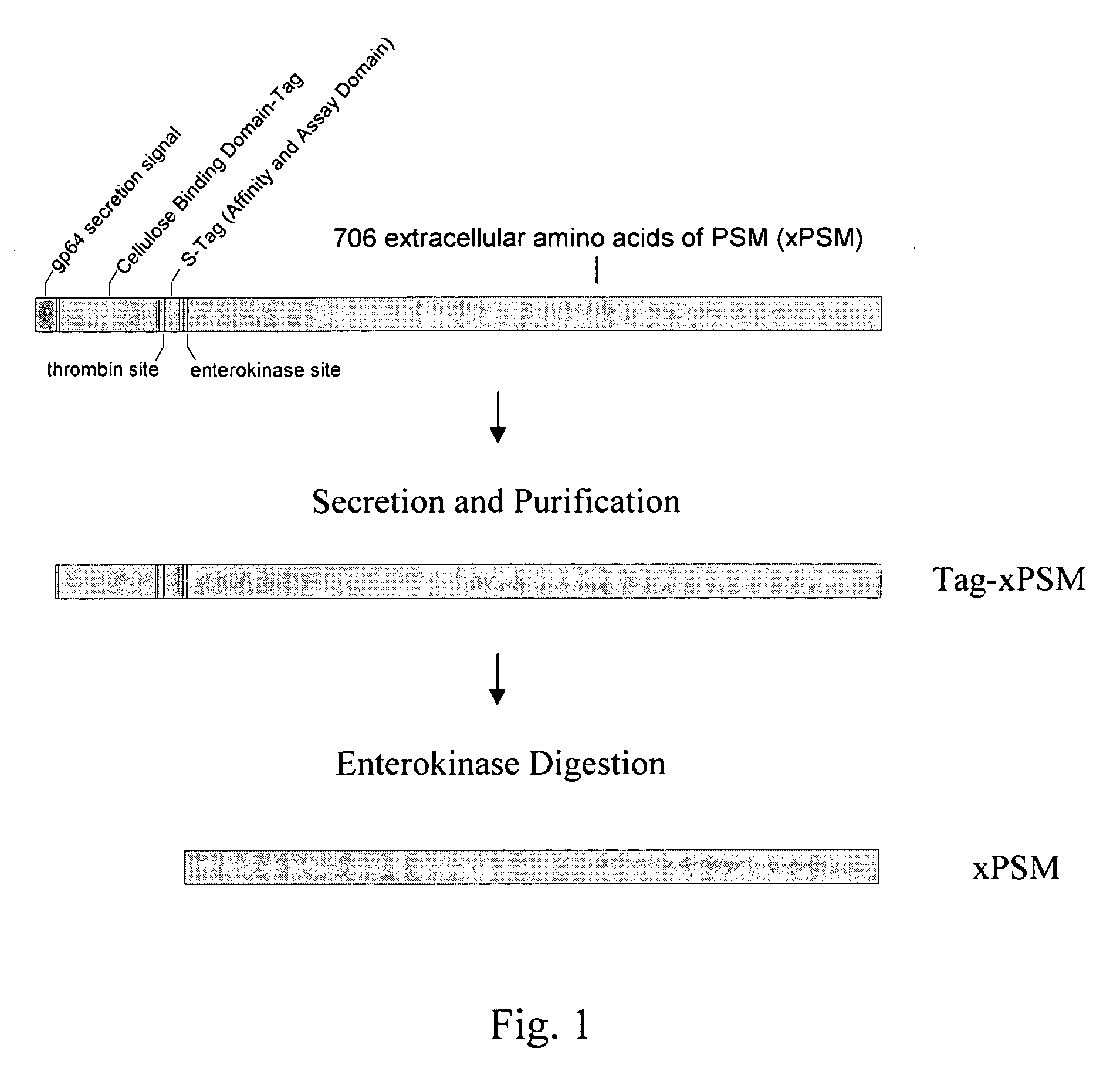
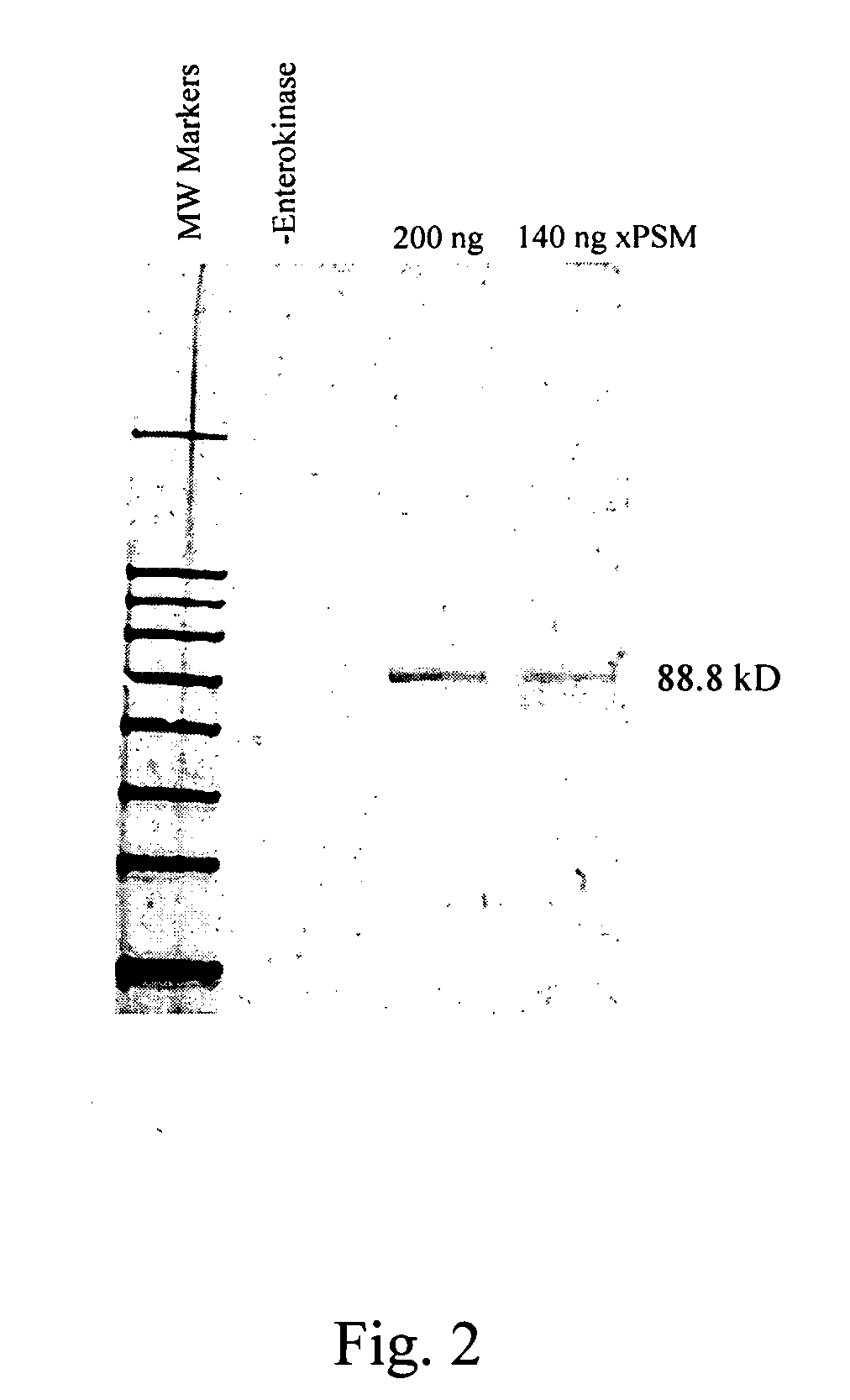
Nucleic acid ligands to the prostate specific membrane antigen
a prostate specific and nucleic acid ligand technology, applied in the direction of group 3/13 element organic compounds, peptides, drug compositions, etc., can solve the problems of limited use of proteins as drugs and reagents, and no reported rna aptamers to membrane bound tumor antigens, etc., to inhibit native psma enzymatic activity and high affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Use of SELEX to Obtain Nucleic Acid Ligands to PSMA
Materials and Methods
[0087] Cloning PSMA cDNA from LNCaP.
[0088] First strand cDNA was synthesized from 2 μg total LNCaP RNA using Superscript II RNase H Reverse Transcriptase (Life Technologies, Inc). Primers homologous to PSMA cDNA bases 134-152 and 2551-2567, flanking the entire full-length PSMA coding region, were used for PCR amplification. Amplification was performed using high fidelity Pfu DNA Polymerase (Stratagene, La Jolla, Calif.). The isolated product was ligated into the pCR-2.1 vector (Invitrogen Corporation, Carlsbad, Calif.). One successful clone, pFULPSM-1, was sequenced and found identical to Genbank accession number M99487. This clone represents the coding region for the full length PSMA protein
[0089] Preparation of Recombinant PSMA Expressing Baculovirus.
[0090] Primers containing restriction enzyme cut sites were designed to overlap the sequence of the entire extracellular portion of PSMA plus a linking glyc...
example 2
Determination of Minimal Size of Aptamers A10 and A9
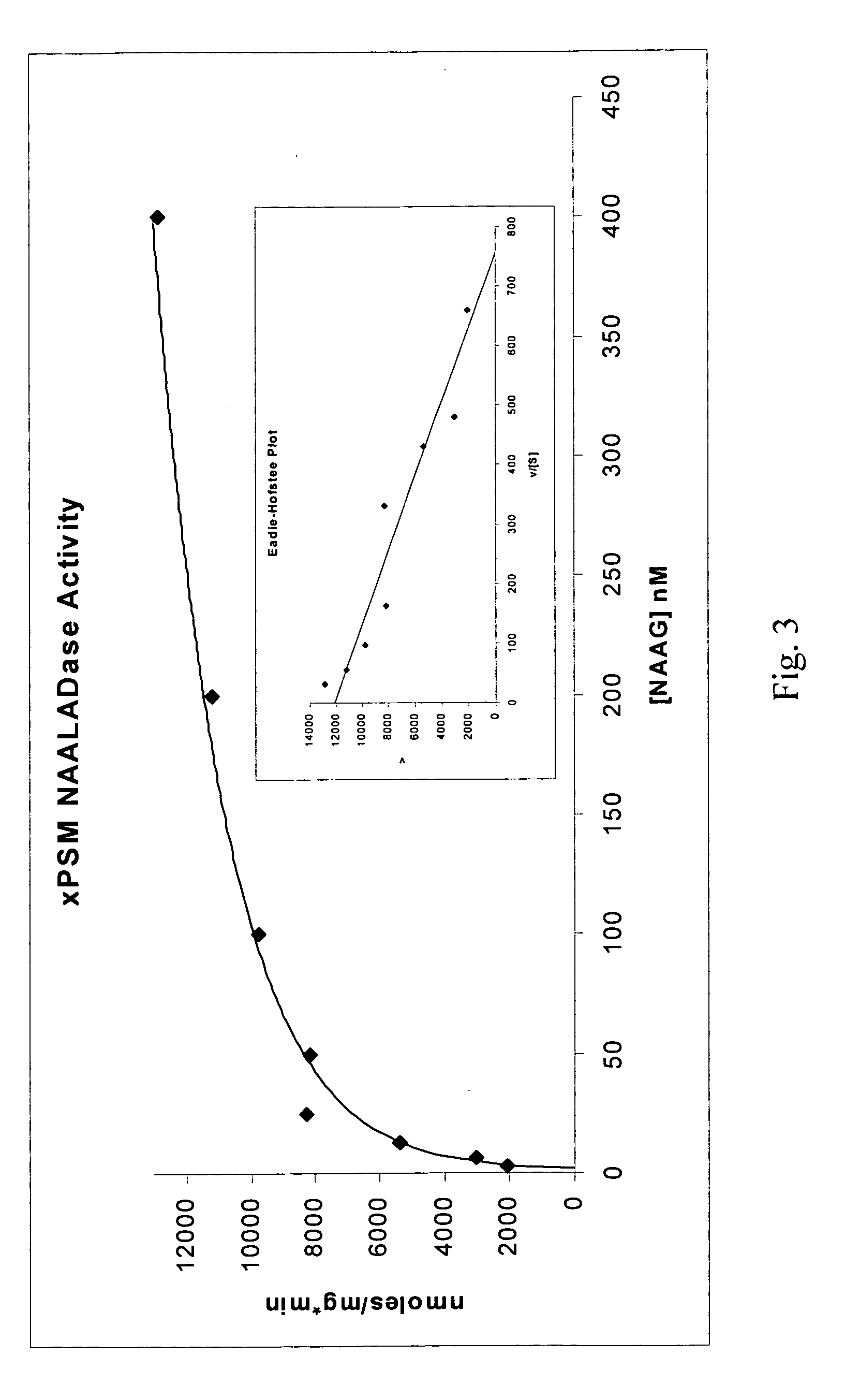
[0112] To determine minimal aptamer sequences, a series of 3′ and 5′ truncations were tested for IC50. Five nucleotides could be removed from the 3′ end of aptamer xPSM-A9 (SEQ ID NO:5) with retention of activity, yielding aptamer A9-1 (SEQ ID NO:6). It was found that at least 15 nucleotides could be deleted from the 3′ end of aptamer xPSM-A10 (SEQ ID NO:15) with retention of activity, yielding aptamer A10-3 (SEQ ID NO:18). This 18.5 kD aptamer retains the ability to inhibit xPSM NAALADase activity with a KI of 20.5 nM. (FIG. 8). The shorter A10-3 could not survive 5′-truncation.
TABLE 1RoundSignal / Noise13.4226.8315.8425.75125.065688.0
[0113]
TABLE 2Reaction mixtures for reverse transcription (RT), QPCR and Transcription5X RT mixQPCR mixTranscription mix2.5 μL 10X RT buffer 10 μL 10X SQ buffer40 μL 5X nucleotides 1 μL 25 mM dNTP′S 1 μL 3′ primer 100 μM40 μL 5X Ribomax 1 μL H2O0.5 μL 5′ primer 100 μM10 μL guanosine 100 mM0.5 μL A...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com