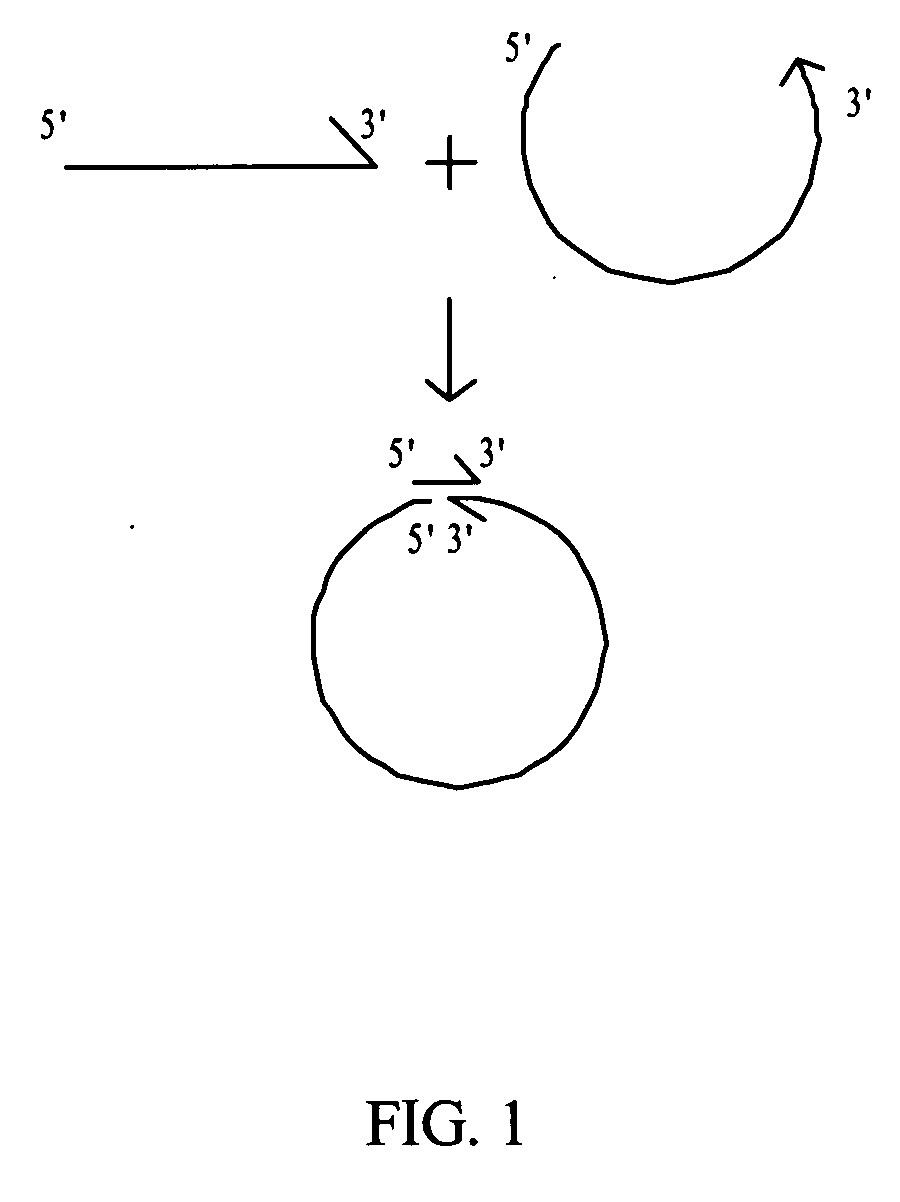
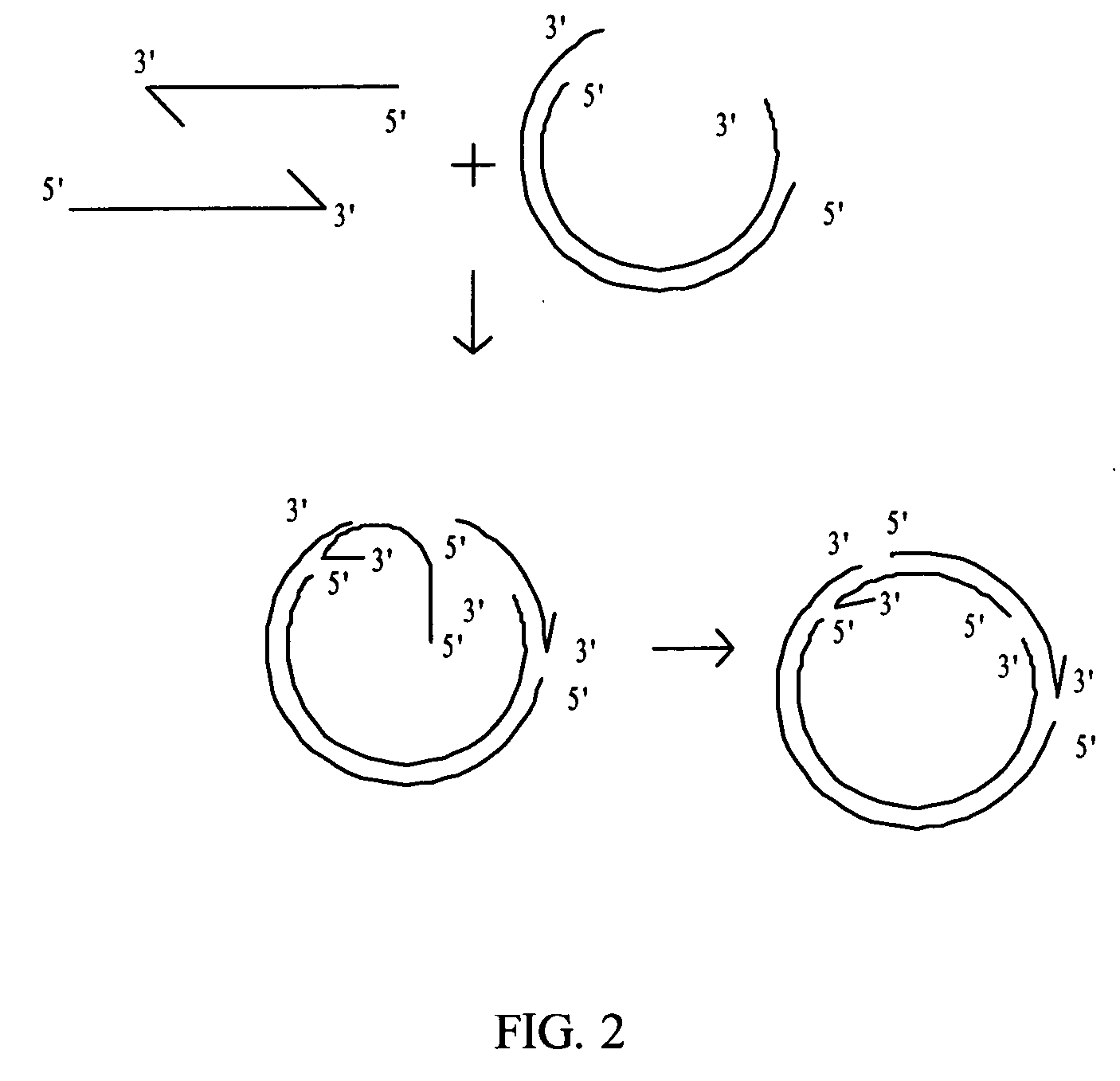
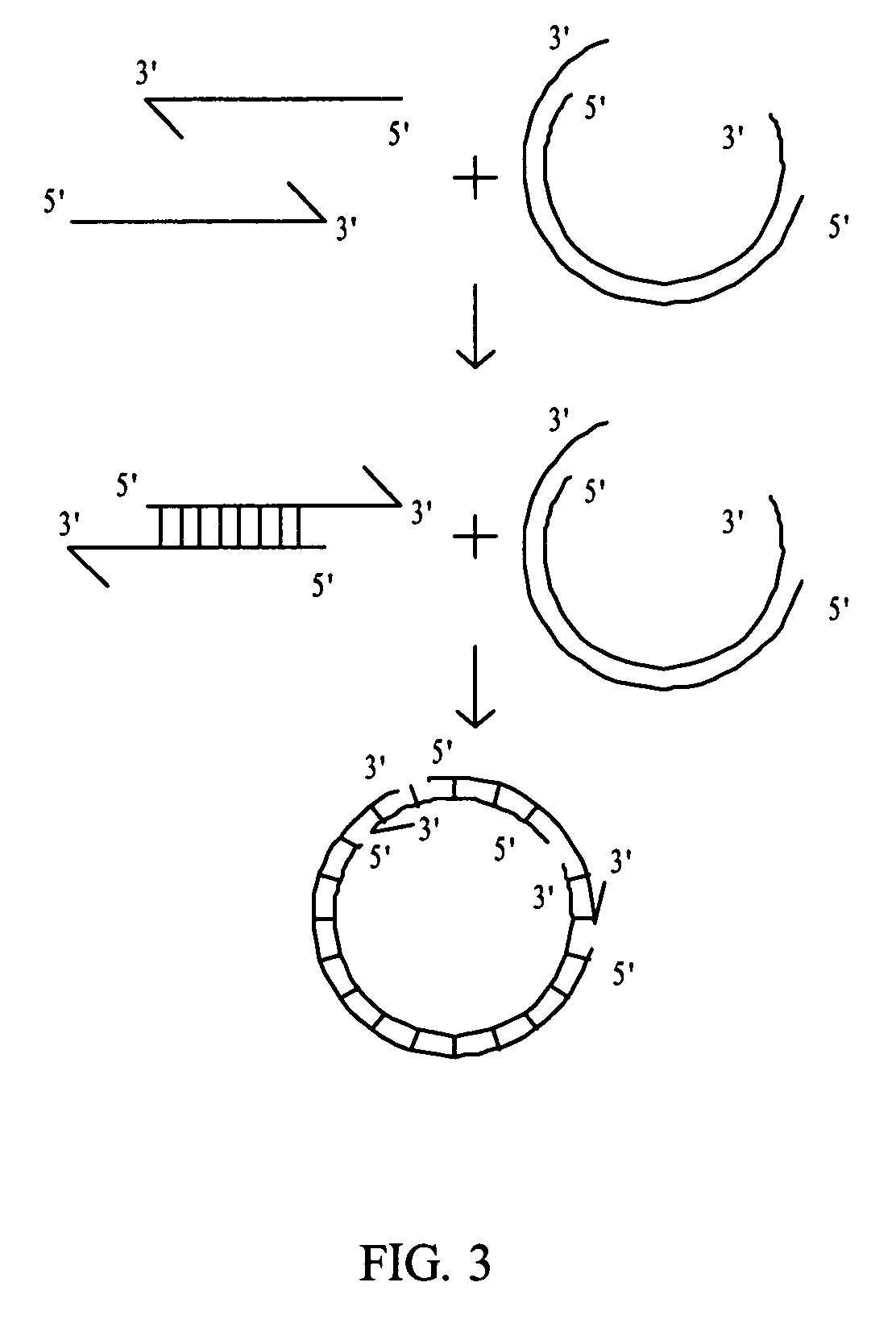
Rolling circle amplification of micro-RNA samples
a micro-rna and rna technology, applied in the field of molecular biology, can solve the problems of affecting the relative proportions of nucleotide sequences, and gc-rich sequences, and achieve the effect of under-representation of the 5′ ends of products, and poor amplification of pcr nucleotide sequences in a prior cycl
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesis of Double-Strand cDNA
[0107] Total RNA was isolated using the Nanoprep™ kit (Stratagene). The (dT)-T7 primer (SEQ ID NO:1) was HPLC purified then treated with kinase. The (dT)-T7 primer was incubated with 100 pg total RNA. A reverse transcription reaction was performed by the addition of 5× First Strand Buffer, 100 mM DTT, 10 mM dNTP, T4gp32, RNase Inhibitor, and Superscript II reverse transcriptase. The dNTP solution contained equimolar amounts of the four nucleotides. The reaction mixture was incubated. Second strand synthesis was performed by the addition of 5× Second Strand Buffer, 10 mM dNTP, DNA Polymerase I, 1 U E. coli RNAse H, E. coli DNA ligase, and T4 DNA polymerase. The reaction mixture was incubated. The reaction mixture was treated with Exonuclease I. The double-stranded cDNA was purified using the Microcon YM-100 system from Millipore.
example 2
Addition of polyA Tails by Terminal Transferase
[0108] 10× buffer, 25 mM CoCl2, 10 mM dATP, and terminal transferase were added to the cDNA such that the concentration of these components in the terminal transferase reaction was 1× buffer, 2.5 mM CoCl2, and 1.0 mM dATP. The reaction mixtures were incubated. The cDNA was purified using the Microcon YM-100 system from Millipore.
example 3
Circularization of the cDNA by a Splint Oligonucleotide
[0109] T tail primer (SEQ ID NO:2), a splint oligonucleotide, was incubated with kinase. The T tail primer annealed to the polyA tailed, double-stranded cDNA. 10× buffer, 1.0 μl 1 mM dTTP, Klenow fragment, and T4 ligase were added to the reaction components. The reactions were incubated. The cDNA was purified using the Microcon YM-100 system from Millipore.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Homogeneity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com