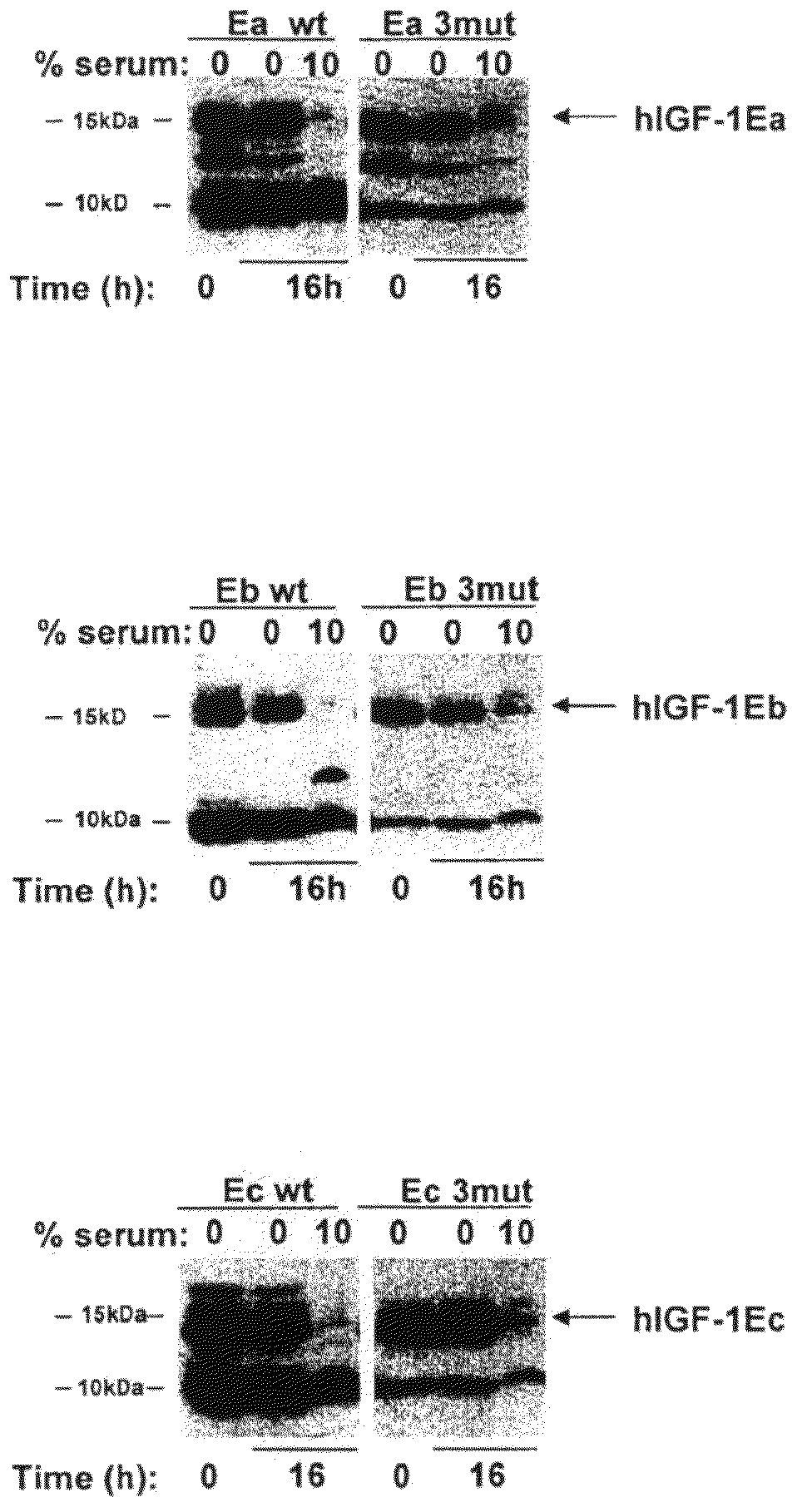Stabilized insulin-like growth factor polypeptides
a growth factor and polypeptide technology, applied in the direction of peptide/protein ingredients, extracellular fluid disorder, metabolism disorder, etc., can solve the problems that igf-1 and igf-2 appear to be poor drug candidates, and achieve the effect of avoiding or reducing this cleavag
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
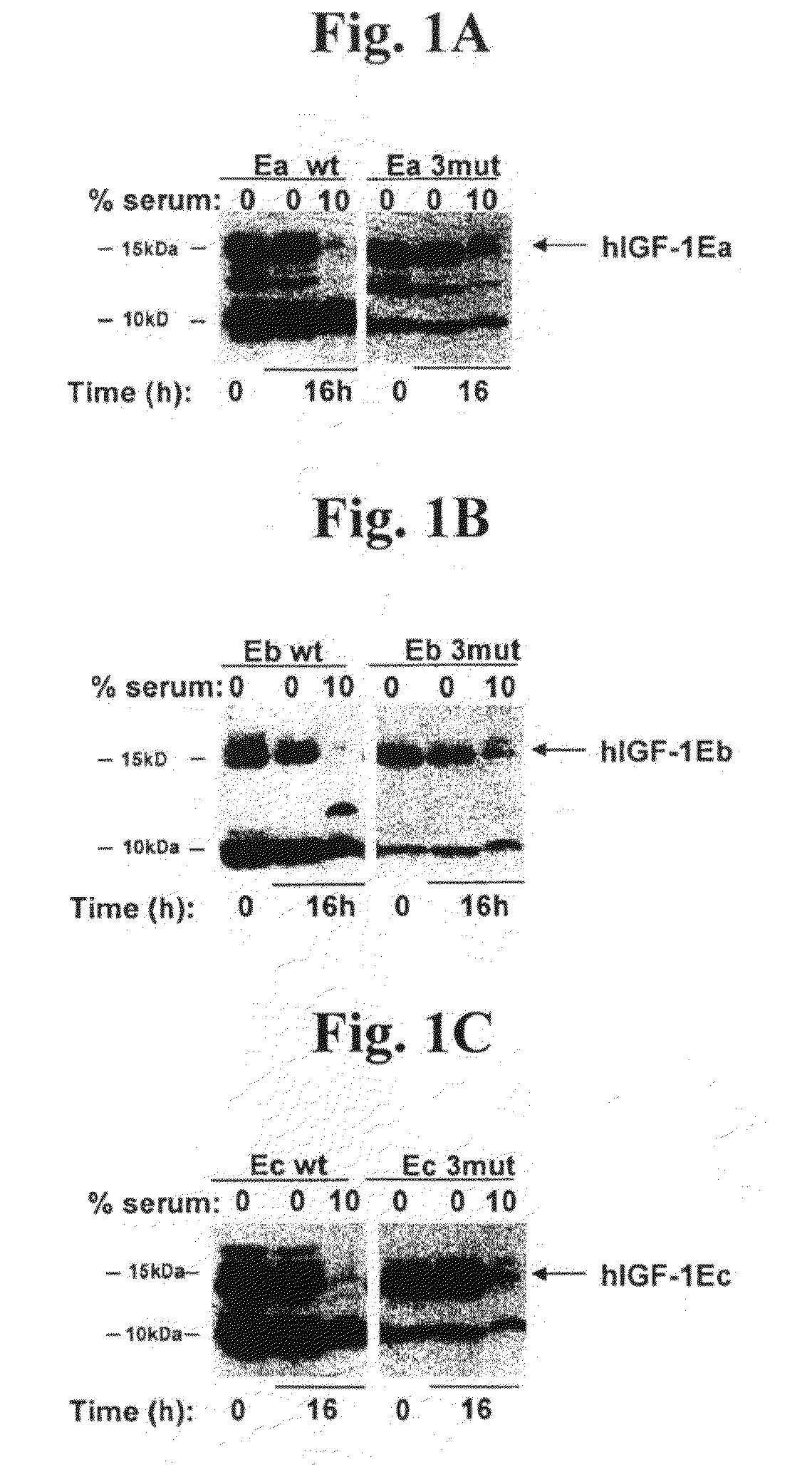
[0111]A DNA expression vector encoding the hIGF-1-Ea precursor polypeptide containing the following modifications was constructed: deletion of G1, deletion of P2, and deletion of E3; mutation of R37 to A; and deletion of R71 and deletion of S72. These mutations are sometimes referred to as “3mut” throughout the present disclosure. This results in the following secreted protein sequence:
(SEQ ID NO: 8)tlcgaelvdalqfvcgdrgfyfnkptgygsssraapqtgivdeccfrscdlrrlemycaplkpaksavraqrhtdmpktqkevhlknasrgsagnknyrm
[0112]Cos7 cells (available from ATCC) were maintained in DMEM containing 10% fetal bovine serum, 100 U / ml penicillin, and 100 μg / ml streptomycin and plated at a density of 1×106 cells per 10-cm plate. These cell cultures were transfected with 8 μg of expression plasmid using Fugene (Roche) according to manufacturer's instructions. Twenty-four hours post-transfection, cells were washed once and cultured in serum-free medium for 48 hours. Supernatants were collected and stored at −80° C.
[01...
example 2
[0116]A DNA expression vector encoding the hIGF-1-Eb precursor polypeptide containing the following mutations was constructed: deletion of G1, deletion of P2, and deletion of E3; mutation of R37 to A; and deletion of R71 and deletion of S72 (i.e., the “3mut”). This results in the following secreted protein sequence:
(SEQ ID NO: 9)tlcgaelvdalqfvcgdrgfyfnkptgygsssarapqtgivdeccfrscdlrrlemycaplkpaksavraqrhtdmpktqkyqppstnkntksqrrkgwpkthpggeqkegteaslqirgkkkeqrreigsrnaecrgkkgk
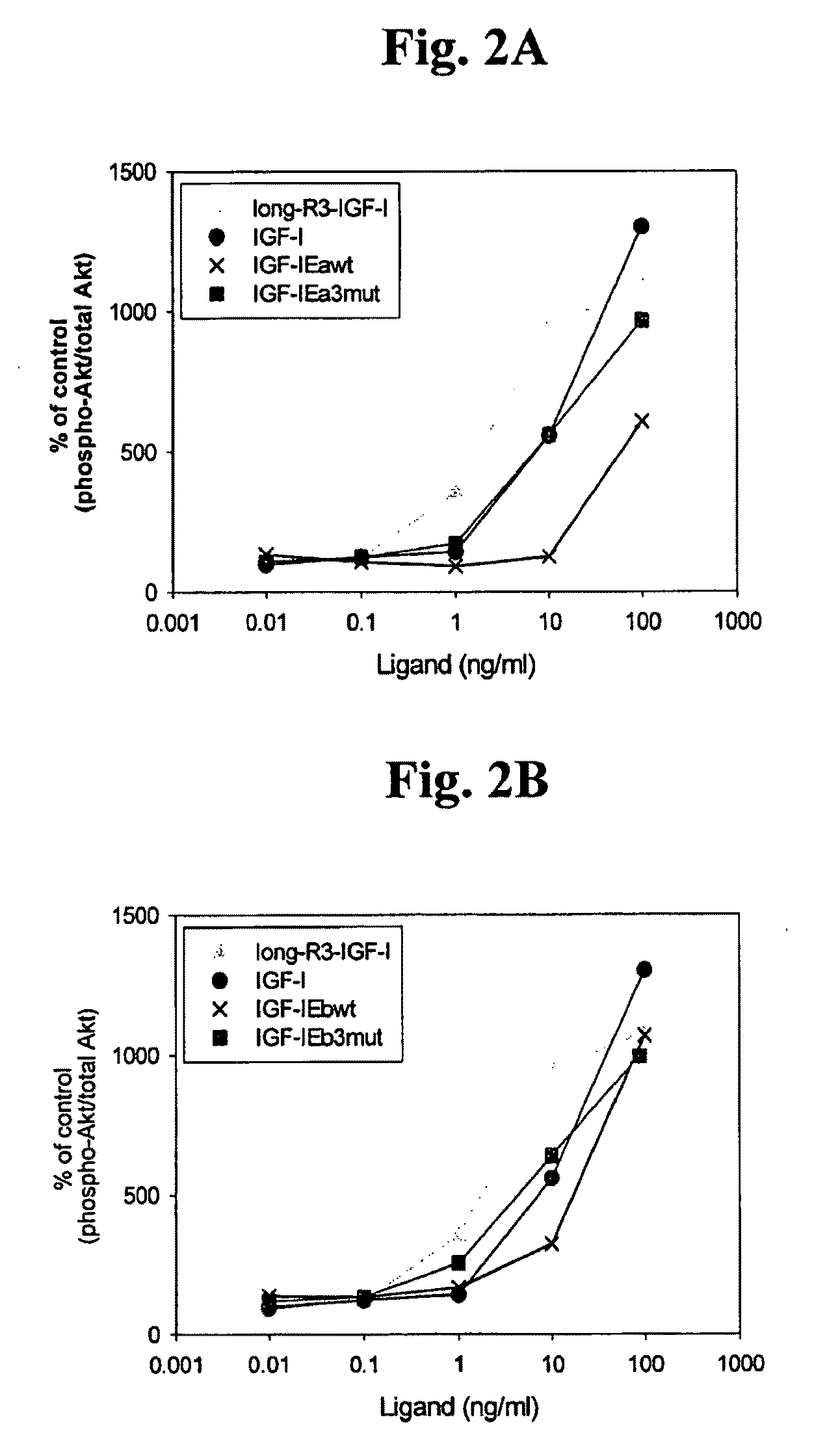
[0117]The polypeptide was assayed in accordance with the procedures described in Example 1 above. FIG. 1B and use of densitometry indicated that the ratio of uncleaved to cleaved IGF-1 was about 1:9, while the ratio for hIGF-1Eb3mut was about 1:1, showing that these modifications result in a stabilized polypeptide. FIG. 2B indicates that the hIGF-1Eb3mut was able to activate the IGF-1R cellular pathway to a similar extent as the long-R3-IGF-1 positive control reagent and the recombinant IGF-1. In addition, the data in ...
example 3
[0118]A DNA expression vector encoding the hIGF-1-Ec precursor polypeptide containing the following mutations was constructed: deletion of G1, deletion of P2, and deletion of E3; mutation of R37 to A; and deletion of R71 and deletion of S72 (i.e., the “3mut”). This results in the following secreted protein sequence:
(SEQ ID NO: 10)tlcgaelvdalqfvcgdrgfyfnkptgygsssrapqtgivdeccfrscdlrrlemycaplkpaksavraqrhtdmpktqkyqppstnkntksqrrkgstfeerk
[0119]The polypeptide was assayed in accordance with the procedures described in Example 1 above. FIG. 1C and use of densitometry indicated that the ratio of uncleaved to cleaved IGF-1 was about 1:5, while the ratio for hIGF-1Ec3mut was about 1:0.96, showing that these modifications result in a stabilized polypeptide. FIG. 2D indicates that the hIGF-1Ec3mut was able to activate the IGF-1R cellular pathway to a similar extent as the long-R3-IGF-1 positive control reagent and the recombinant IGF-1. In addition, the data in FIG. 5 directly shows that hIGF-1E...
PUM
| Property | Measurement | Unit |
|---|---|---|
| body weight | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
| Ea | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com