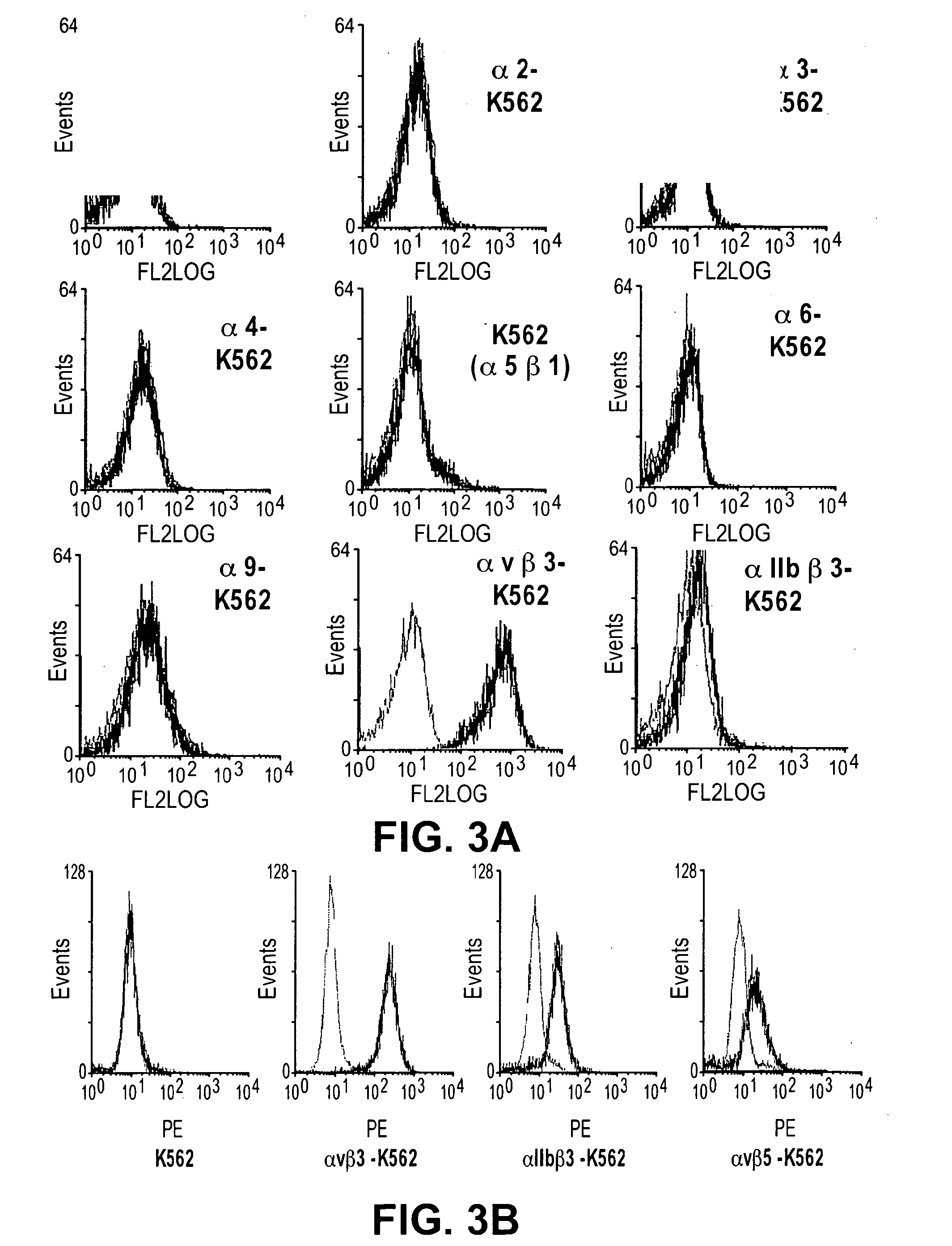
Rgd-containing cyclic peptides
a technology of cyclic peptides and peptides, which is applied in the direction of peptides, peptide/protein ingredients, immunoglobulins, etc., can solve the problem of limited number of peptides that can be simultaneously tested in the “spatial screening” process, and achieve the effect of inhibiting the activity of v3 integrin
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of 8Mer Library 1
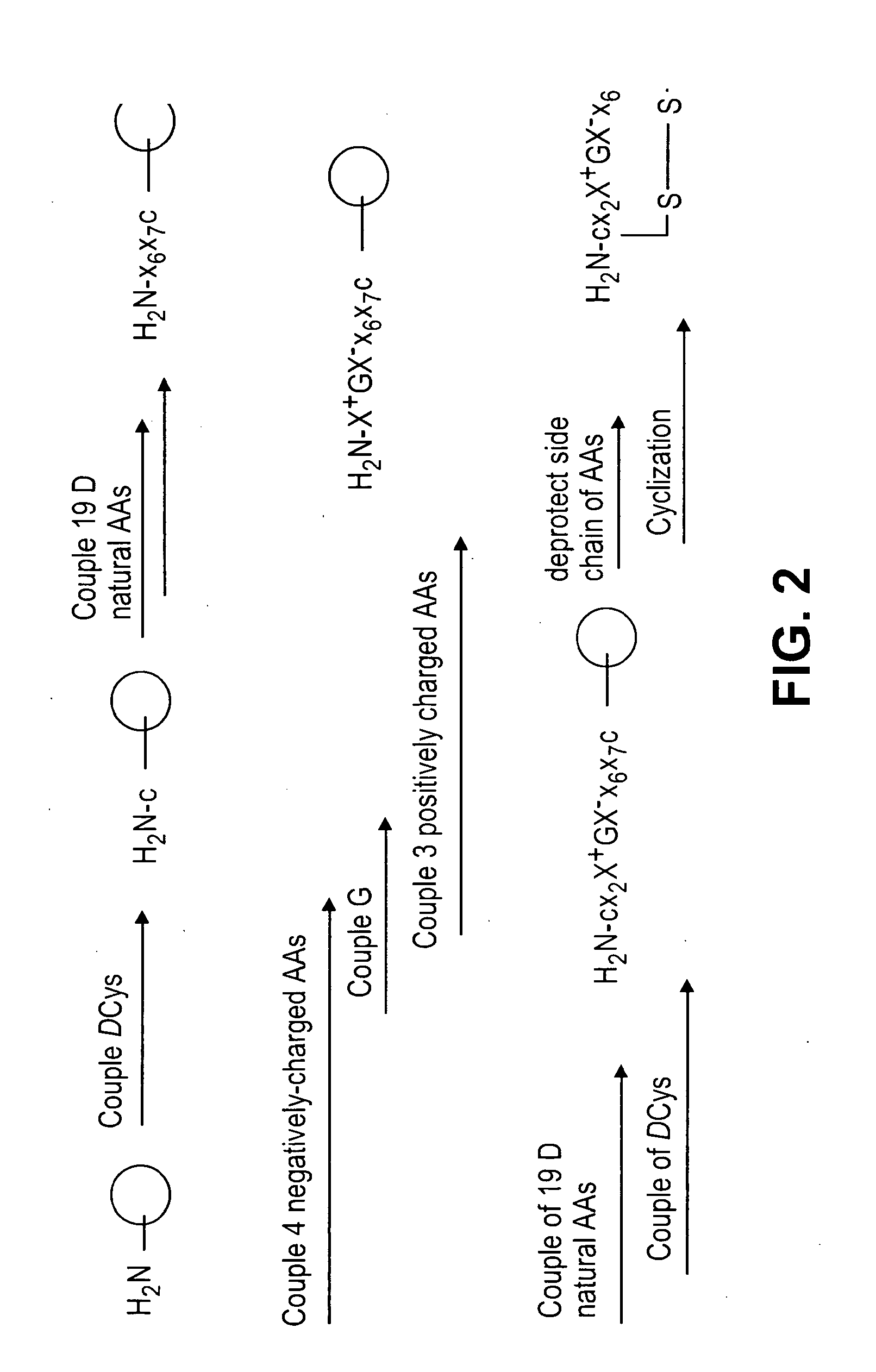
[0116]Library 1 was prepared according to the scheme shown in FIG. 2. TentaGel resin (2 g, loading: 0.27 mmole / g) was swollen in DMF for 4 h. After filtration, a solution of Fmoc-D-Cys(Trt)-OH (948.83 mg, 1.62 mmole), HOBt (218.7 mg, 1.62 mmole) and DIC (250.85 μL, 1.62 mmole) was added to the beads. The coupling reaction was carried out at room temperature for 2 h. After filtration, the beads were washed three times each with DMF, MeOH and DMF. The beads were subjected to Fmoc deprotection with 20% 4-methyl piperieine (5 min, 15 min). After washing with DMF, MeOH and DMF, the beads were split into 19 columns and respectively coupled with 19 natural D amino acid (See Table 3) in a similar manner described above. According to the standard “split-mix” approach, the library was completed. The beads were then dried under vacuum for overnight before adding a TFA-based cleavage cocktail (TFA: phenol: water: thioanisole: Tis, 10:0.5:0.5:0.5:0.25, v / w / v / v / v) for...
example 2
Preparation of 9Mer Library 2
[0117]The same synthetic route and amino acids used in the synthesis of library 1, were employed with the exception of an additional amino acid added at x8 position to create a 9mer.
example 3
Preparation of 8Mer Library 3
[0118]Library 3 was prepared according to the scheme shown in FIG. 10. TentaGel resin (2 g, loading: 0.27 mmole / g) was swollen in water for 24 h. After filtration, a solution of Fmoc-OSu (7.29 mg, 0.0216 mmole), Alloc-OSu (17.2 mg, 0.0864 mmole) and DIPEA (37.60 μL, 0.216 mmole) in DCM / diethyl ether (110 mL: 90 mL) was added to the beads, and the reaction was allowed to occur under vigorously shaking for 30 min. Then the inner layer of the beads was protected by treatment with (Boc)2O (589.30 mg, 2.7 mmole) / DIPEA (940.4 μL, 5.4 mmole) for 2 h. After Alloc deprotection, a solution of Ac-Gly-OH (101.12 mg, 0.864 mmole), HOBt (117 mg, 0.864 mmole) and DIC (128.3 μL, 0.864 mmole) in DMF was used to block the exposed N-terminus on the bead surface. Upon Fmoc deprotection and Boc deprotection, the library was synthesized under the same condition as library 1, following the standard “split-and-mix” approach. The beads were then dried under vacuum overnight befo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com