Recombinant production of steviol glycosides
a technology of steviol glycosides and recombinant production, which is applied in the direction of transferases, baking gum, bakery products, etc., can solve the problems of undesirable off-flavors of contaminants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Selection of Candidate UGT Genes
[0135]Phylogenetic and protein BLAST analysis were used to identify 7 candidate genes belonging to the UGT91 subfamily for 1,2-19-O-glucose glycosylation activity (Table 1).
TABLE 1List of UGT candidate genesNameDescriptionAccessionSequence IDBD1PREDICTED: UDP-XP_003560664.1SEQ ID NO: 1glycosyltransferase 91C1-like[Brachypodium distachyon]BD2PREDICTED: UDP-XP_003560669.1SEQ ID NO: 2glycosyltransferase 91C1-like[Brachypodium distachyon]BD3PREDICTED: LOWXP_003581636.1SEQ ID NO: 3QUALITY PROTEIN: UDP-glycosyltransferase 91C1-like[Brachypodium distachyon]BD4PREDICTED: UDP-XP_003580515.1SEQ ID NO: 4glycosyltransferase 91C1-like[Brachypodium distachyon]BD5PREDICTED: LOWXP_003559500.1SEQ ID NO: 5QUALITY PROTEIN: UDP-glycosyltransferase 91B1-like[Brachypodium distachyon]HV1predicted protein [HordeumBAJ98242.1SEQ ID NO: 6vulgare subsp. vulgare]HV2predicted protein [HordeumBAJ93155.1SEQ ID NO: 8vulgare subsp. vulgare]
example 2
Enzymatic Activity Screening of Candidate UGT Genes
[0136]Full length DNA fragments of all candidate UGT genes were commercially synthesized. Almost all codons of the cDNA were changed to those preferred for E. coli (Genscript, NJ). The synthesized DNA was cloned into a bacterial expression vector pETite N-His SUMO Kan Vector (Lucigen).
[0137]Each expression construct was transformed into E. coli BL21 (DE3), which was subsequently grown in LB media containing 50 μg / mL kanamycin at 37° C. until reaching an OD600 of 0.8-1.0. Protein expression was induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and the culture was further grown at 16° C. for 22 hr. Cells were harvested by centrifugation (3,000×g; 10 min; 4° C.). The cell pellets were collected and were either used immediately or stored at −80° C.
[0138]The cell pellets typically were re-suspended in lysis buffer (50 mM potassium phosphate buffer, pH 7.2, 25 ug / ml lysozyme, 5 ug / ml DNase I, 20 mM imidazole, 500 mM...
example 3
Steviol Glycoside Biosynthesis Using the Recombinant HV1 Polypeptide
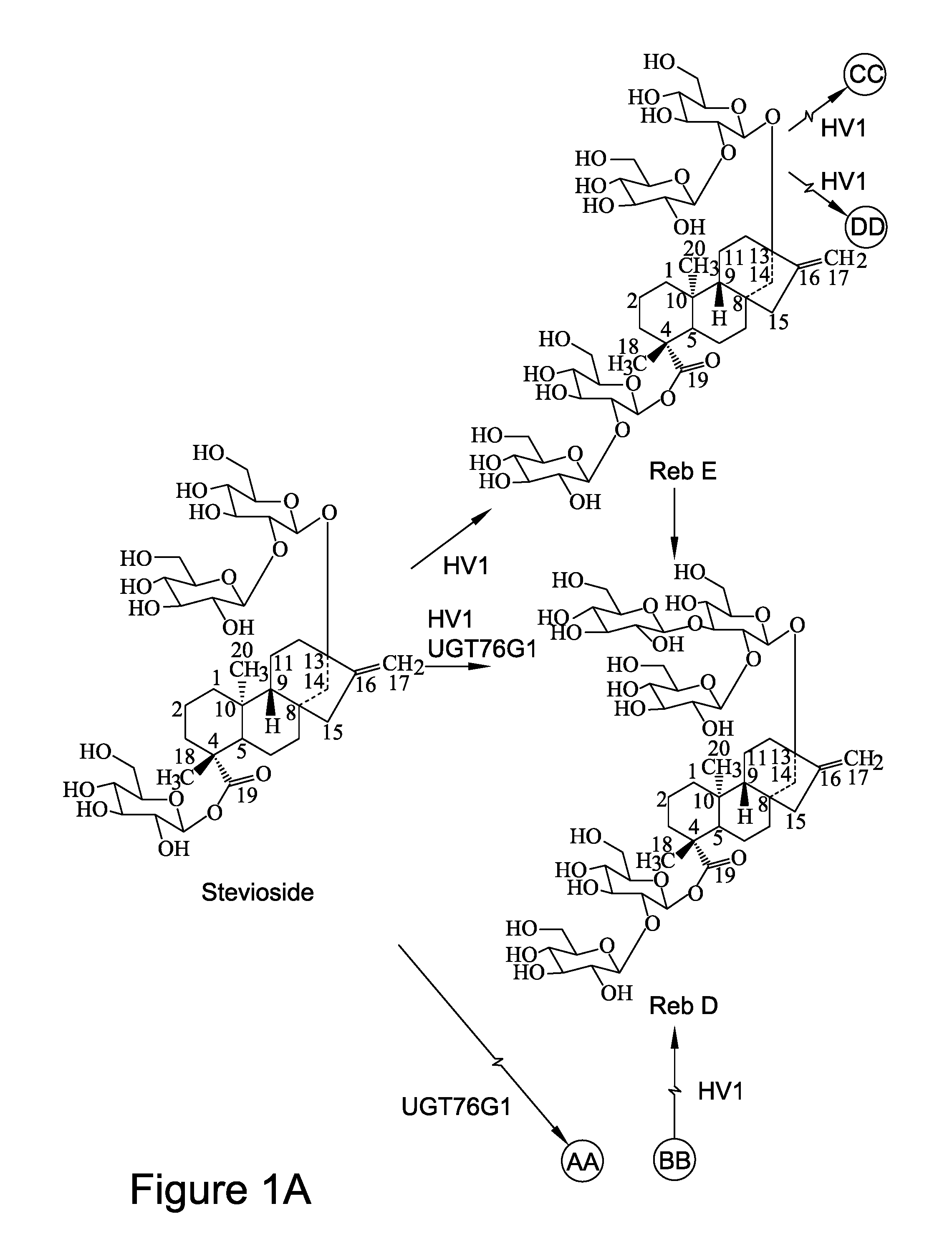
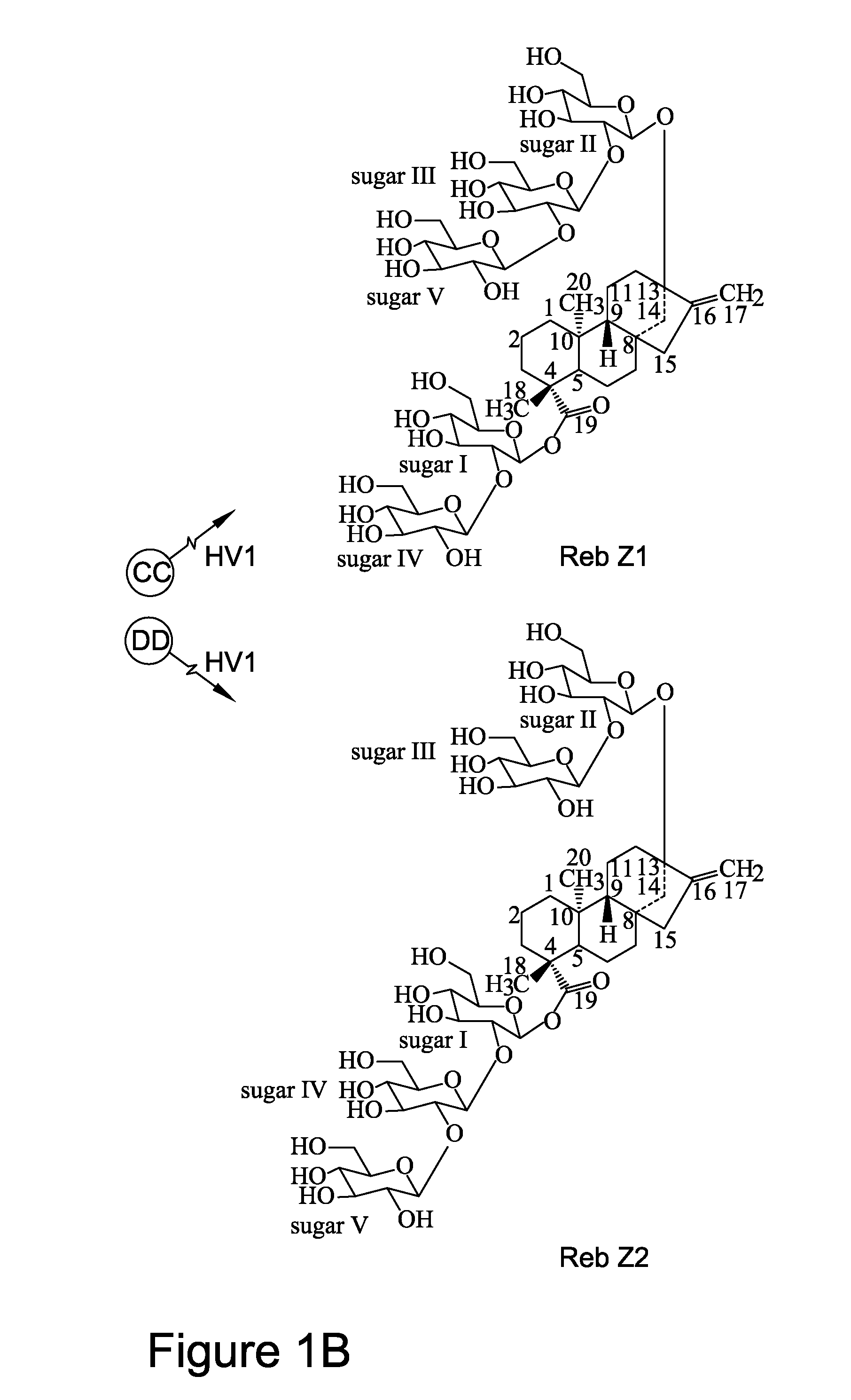
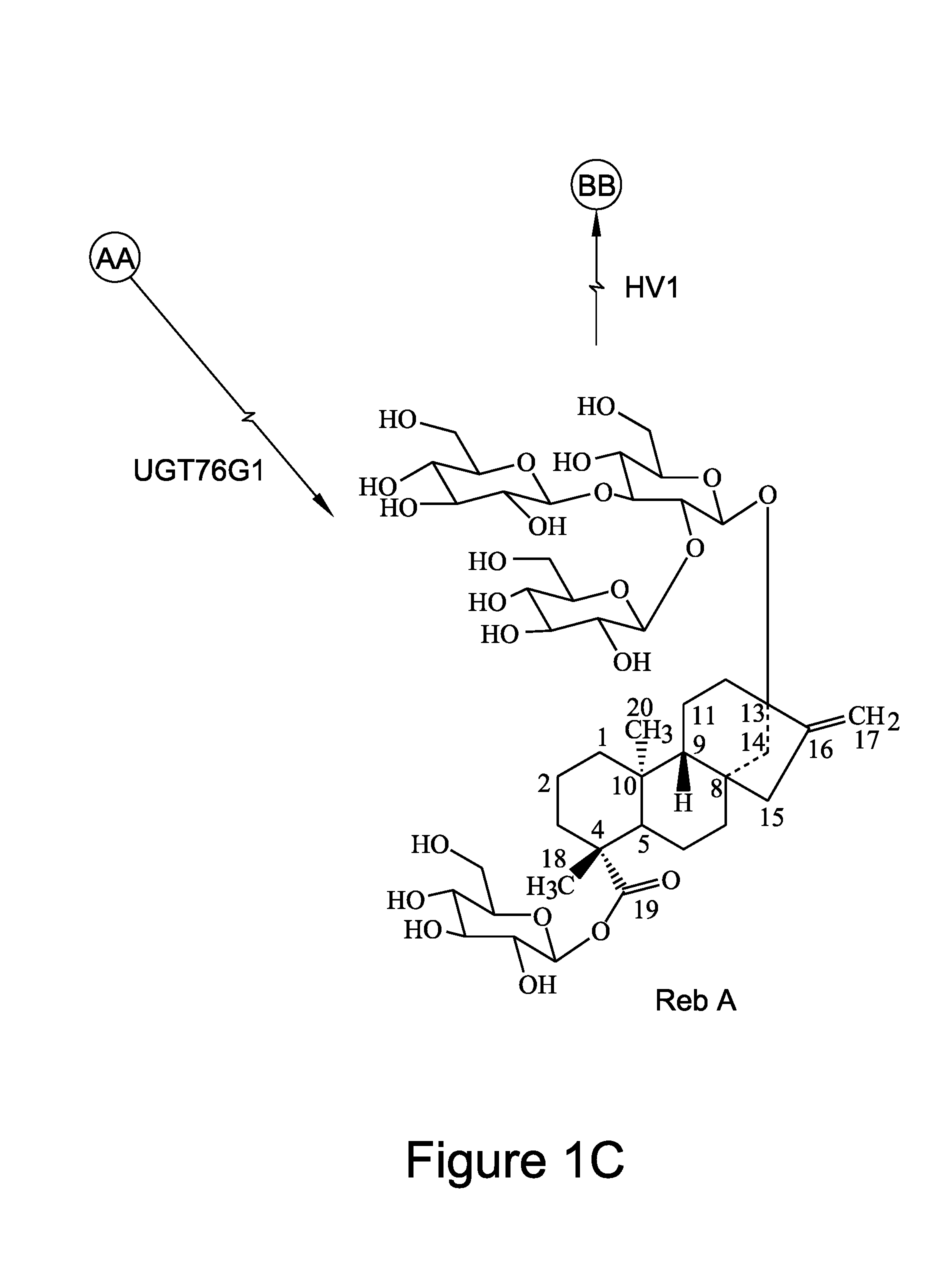
[0146]As shown in FIGS. 1A-1C, rebaudioside D can also be formed by glycosylation of the C-3′ of the C-13-O-glucose of rebaudioside E. Thus, rebaudioside D can be produced by different biosynthetic routes (e.g. via rebaudioside A vs. rebaudioside E), depending on the orders in which the glycosylation reactions occur. For example, glycosylation at C-3′ of the C-13-O-glucose of stevioside can occur first to produce the intermediate rebaudioside A, followed by glycosylation at C-2′ of the 19-O-glucose of rebaudioside A to produce rebaudioside D. So far, UGT76G1 (SEQ ID NO:11) from stevia has been identified as an enzyme that transfers a sugar residue to C-3′ of the C-13-O-glucose of stevioside to form rebaudioside A.
[0147]Codon optimized UGT76G1 cDNA was inserted in a bacterial expression vector, and the recombinant UGT76G1 protein was expressed and purified by affinity chromatography. The purified recombinant UGT76G1 ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com