Use of lifr or fgfr3 as a cell surface marker for isolating human cardiac ventricular progenitor cells
a cell surface marker and progenitor cell technology, applied in 3d culture, skeletal/connective tissue cells, instruments, etc., can solve the problems of unsuitable for isolating large quantities of viable cells, unmet clinical needs, etc., to enhance ventricular function, improve function in a damaged heart, and easy and rapid cell isolation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
n of Human Isl1+ Cardiomyogenic Progenitor Cells by Modulation of Wnt Signaling in Human Pluripotent Stem Cells
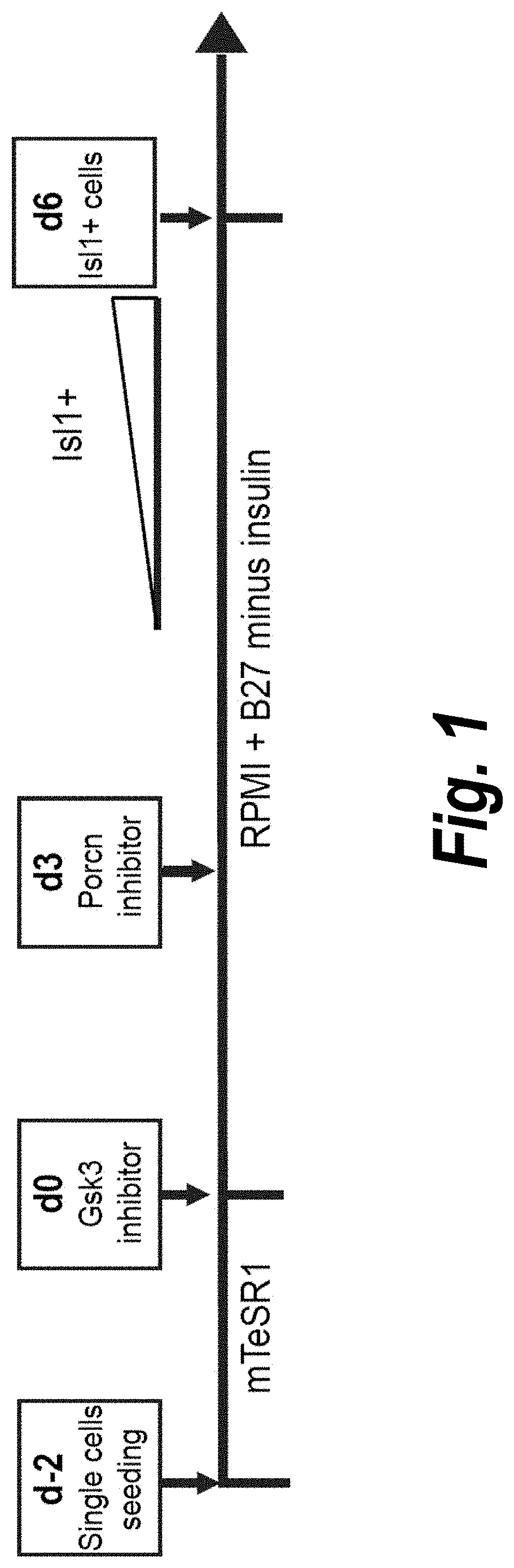
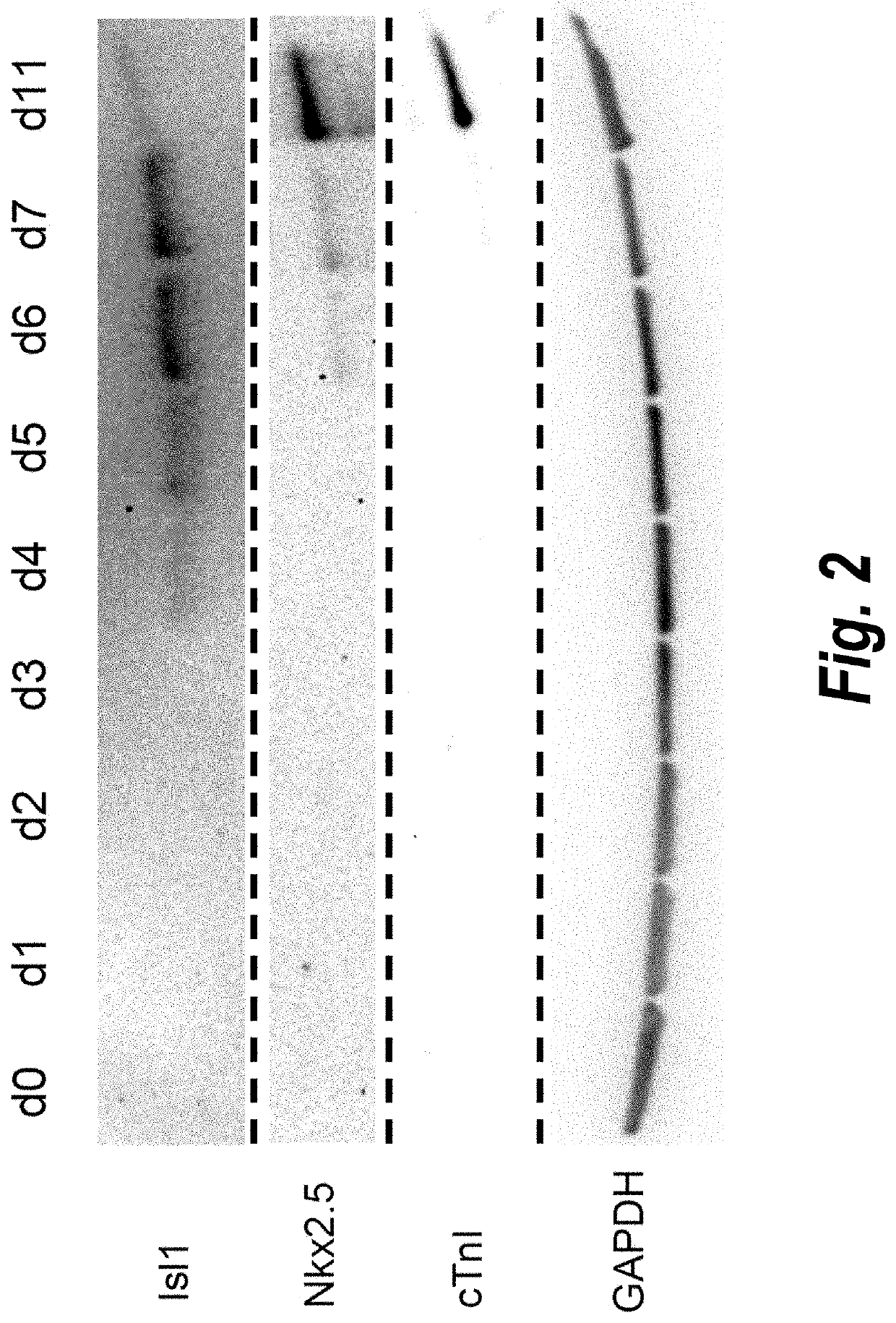
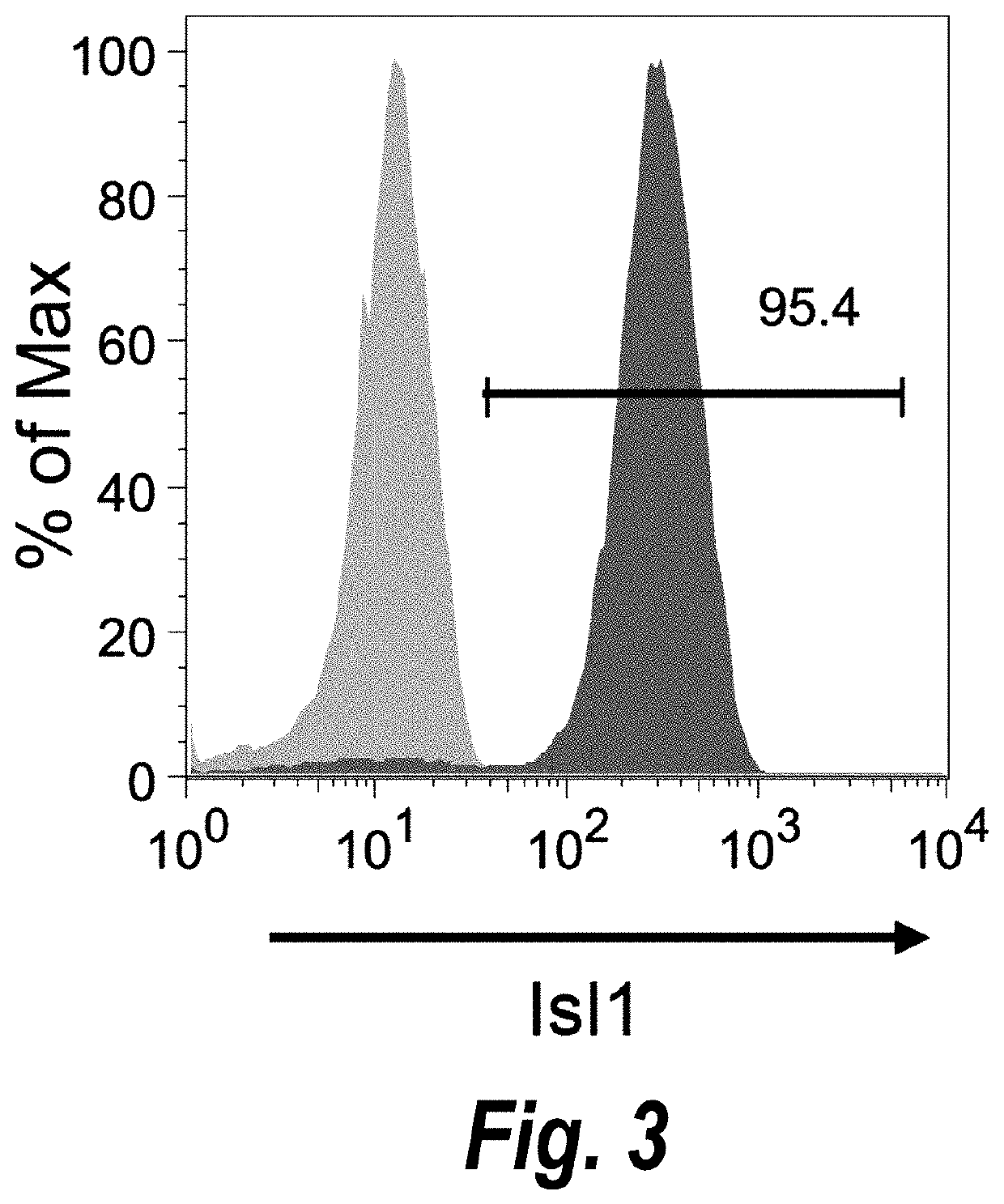
[0207]Temporal modulation of canonical Wnt signaling has been shown to be sufficient to generate functional cardiomyocytes at high yield and purity from numerous hPSC lines (Lian, X. et al. (2012) Proc. Natl. Acad. Sci. USA 109:E1848-1857; Lian, X. et al. (2013) Nat. Protoc. 8:162-175). In this approach, Wnt / β-catenin signaling first is activated in the hPSCs, followed by an incubation period, followed by inhibition of Wnt / β-catenin signaling. In the originally published protocol, Wnt / β-catenin signaling activation was achieved by incubation with the Gsk3 inhibitor CHIR99021 (GSK-3 α, IC50=10 nM; GSK-3, β IC50=6.7 nM) and Wnt / β-catenin signaling inhibition was achieved by incubation with the Porcn inhibitor IWP2 (IC50=27 nM). Because we used Gsk3 inhibitor and Wnt production inhibitor for cardiac differentiation, this protocol was termed GiWi protocol. To improve the effici...
example 2
ation of Jagged 1 as a Cell Surface Marker of Cardiac Progenitor Cells
[0215]To profile the transcriptional changes that occur during the cardiac differentiation process at a genome-scale level, RNA sequencing (RNA-seq) was performed at different time points following differentiation to build cardiac development transcriptional landscapes. We performed RNA-seq experiments on day 0 to day 7 samples, as well as day 19 and day 35 samples (two independent biological replicates per time point). Two batches of RNA-seq (100 bp and 50 bp read length) were performed using the illumine Hiseq 2000 platform. In total, 20 samples were examined. Bowtie and Tophat were used to map our reads into a reference human genome (hg19) and we calculate each gene expression (annotation of the genes according to Refseq) using RPKM method (Reads per kilobase transcript per million reads). Differentiation of hPSCs to cardiomyocytes involves five major cell types: pluripotent stem cells (day 0), mesoderm progeni...
example 3
fferentiation of Isl1+Jag1+ Cardiac Progenitor Cells
[0224]To characterize the clonal differentiation potential of Isl1+Jag1+ cells, cardiomyogenic progenitor cells were generated by the culturing protocol described in Example 1, and one single Isl1+Jag1+ cell was seeded into one well of a Matrigel-coated 48-well plate. Cells were purified with antibody of Jag1 and then one single cell was seeded into one well. The single cells were then cultured for 3 weeks in Cardiac Progenitor Culture (CPC) medium (advanced DMEM / F12 supplemented with 2.5 mM GlutaMAX, 100 μg / ml Vitamin C, 20% Knockout Serum Replacement).
[0225]Immunostaining of the 3-week differentiation cell population was then performed with three antibodies: cardiac troponin 1 (cTn1) for cardiomyocytes, CD144 (VE-cadherin) for endothelial cells and smooth muscle actin (SMA) for smooth muscle cells. The results showed that the single cell-cultured, Isl1+Jag1+ cells gave rise to cTn1 positive and SMA positive cells, but not VE-cadh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com