Mononucleotide polymorphic site of CDC6 gene correlated to primary liver cancer and application thereof
A single nucleotide polymorphism, nucleic acid technology, applied in the detection of the polymorphic site, prevention, auxiliary diagnosis and treatment of primary hepatocellular carcinoma, single nucleotide polymorphic site, to achieve the effect of promoting discovery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1: Collection of blood samples and extraction of genomic DNA
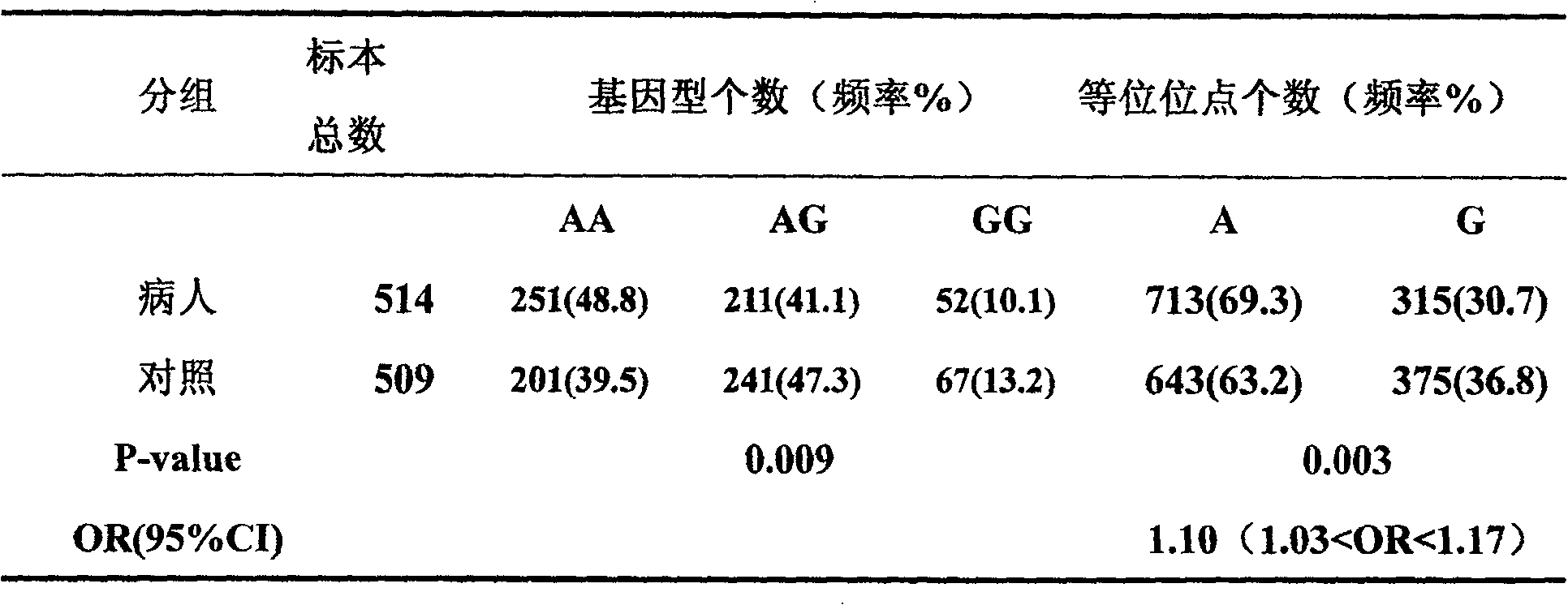
[0037]A total of 514 sporadic primary hepatocellular carcinoma specimens were collected from the Cancer Hospital Affiliated to Sun Yat-sen University in Guangzhou, with an average age of 48.6±11.8 years (standard deviation), and 44 of them were female. All cases were patients who were treated in the cancer hospital, and were diagnosed as primary hepatocellular carcinoma according to the World Health Organization criteria. The control samples were 509 unrelated healthy individuals from Guangdong Province, with an average age of 46.2±14.0 years (standard deviation), including 45 female samples.
[0038] Leukocytes were separated from the blood samples collected above by conventional methods, and genomic DNA of leukocytes was extracted by the phenol-chloroform method, and the sample DNA was uniformly diluted to 10 ng / ul as a DNA template.
Embodiment 2
[0039] Embodiment 2: design nucleotide primer sequence, carry out polymerase chain reaction (PCR) amplification
[0040] The PCR reaction was carried out on MJ PTC-200 PCR instrument.
[0041] The PCR reaction system and procedures are as follows:
[0042] Perform an amplification reaction with a total volume of 20ul for each sample, which contains 20ng of the prepared genomic DNA dilution, 2ul of 10×Taq Buffer, 0.8ul of 5mM dNTP, and 25mM MgCl 2 1.6ul, 0.25 units of Taq DNA polymerase and 0.5ul each of amplification primers (diluted to 10uM), and finally add ddH 2 O make the total volume 20ul. After the reaction was pre-denatured at 94°C for 2 minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, extension at 72°C for 30 seconds, and finally a fill-in at 72°C for 6 minutes were performed.
[0043] Amplification primers are as follows:
[0044] Forward primer: 5'-CTAGGCTTTTCACTGTCACCAA-3' (SEQ ID NO: 2)
[0045] Reverse primer: 5'-G...
Embodiment 3
[0047] Example 3: SNP typing on the amplified DNA sequence
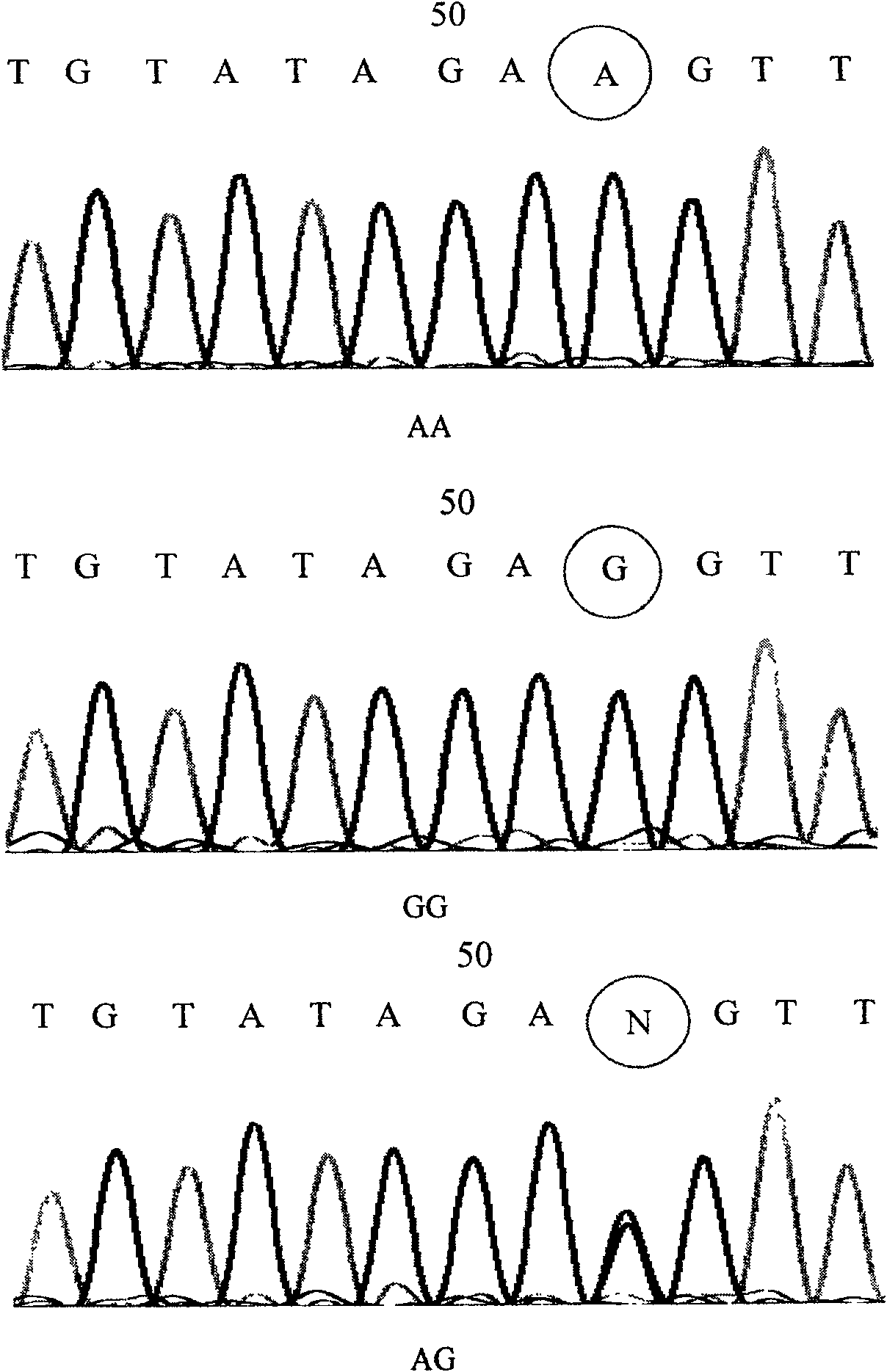
[0048] For the product obtained in Example 2, single-strand conformational polymorphism (SSCP) was used for gene polymorphism typing. The specific method is: add 1 μl PCR product to 4 μl loading buffer (98% deionized formamide, 10 mM EDTA (pH8.0), 0.1% xylene cyanol, 0.1% bromophenol blue), denature at 95°C for 10 minutes, place Place on ice for 2 minutes, quickly spot 4 μl on a 10% non-denaturing polyacrylamide gel (Arc:Bis=37.5:1), and place in a refrigerator at 4°C for electrophoresis. 1.5W constant power electrophoresis for 10 hours, silver staining. The results of SSCP were sequenced and verified, and three genotypes of GG, AG, and AA were found in total. For some sequencing results, see figure 1 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com