Stem-ring type oligonucleotide probe
An oligonucleotide and probe technology, applied in the field of nucleotide structure, can solve the problems of low thermodynamic metastability and high design difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
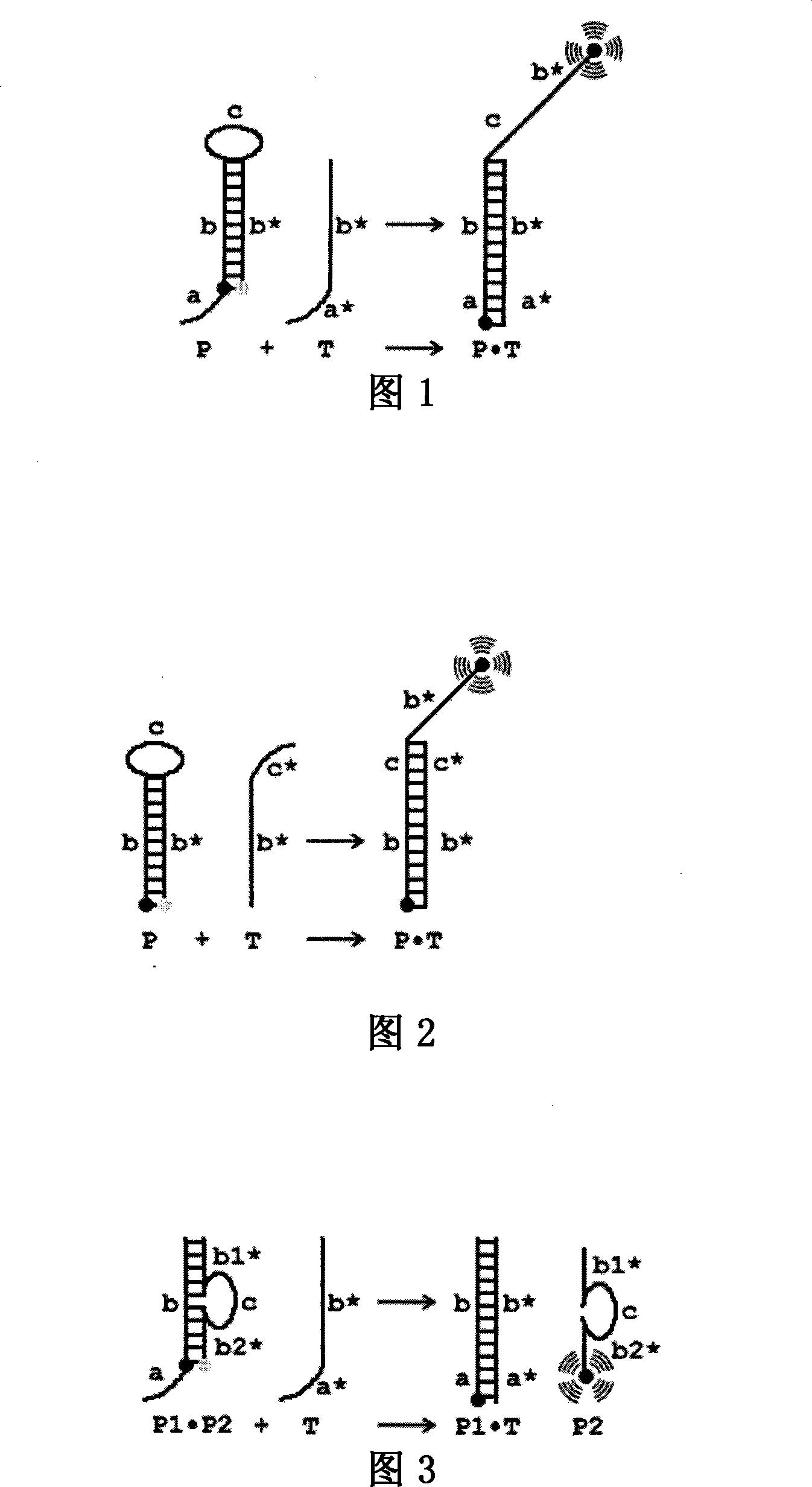
[0078] Example 1. Monomer probes with cohesive ends
[0079] A monomeric oligonucleotide probe P with a stem-loop structure, the probe is composed of an oligonucleotide chain, and its initial state has a loop formed by sequence c, and complementary sequences b and b* combined to form The stem part and the sticky end a are composed of a fluorescent group at the b* end, and a quencher group is marked at the junction of b and a, and the recognition region is composed of the stem b and the sticky end a. The target molecule T is formed by connecting the sequences b* and a*, and is completely complementary to the stem sequence b and the sticky end a of the probe P. When there is no target molecule T in the reaction system, the probe P maintains the initial state, the fluorescence signal of the fluorophore is absorbed by the adjacent quenching group, and there is no detectable fluorescence signal in the system; when there is a target molecule in the reaction system When the molecule...
Embodiment 2
[0080] Example 2. Monomer probes without sticky ends
[0081] A monomeric oligonucleotide probe P with a stem-loop structure, the probe is composed of an oligonucleotide chain, and its initial state has a loop formed by sequence c, and complementary sequences b and b* combined to form The end of the stem is a blunt end, and the two ends are respectively labeled with a fluorescent group and a quencher group. The recognition region is composed of the stem sequence b and the loop sequence c. The target molecule T is formed by connecting the sequences b* and c*, and is completely complementary to the stem sequence b and the loop sequence c of the probe P. When there is no target molecule T in the reaction system, the probe P maintains the initial state, the fluorescence signal of the fluorophore is absorbed by the adjacent quenching group, and there is no detectable fluorescence signal in the system; when there is a target molecule in the reaction system When the molecule is T, t...
Embodiment 3
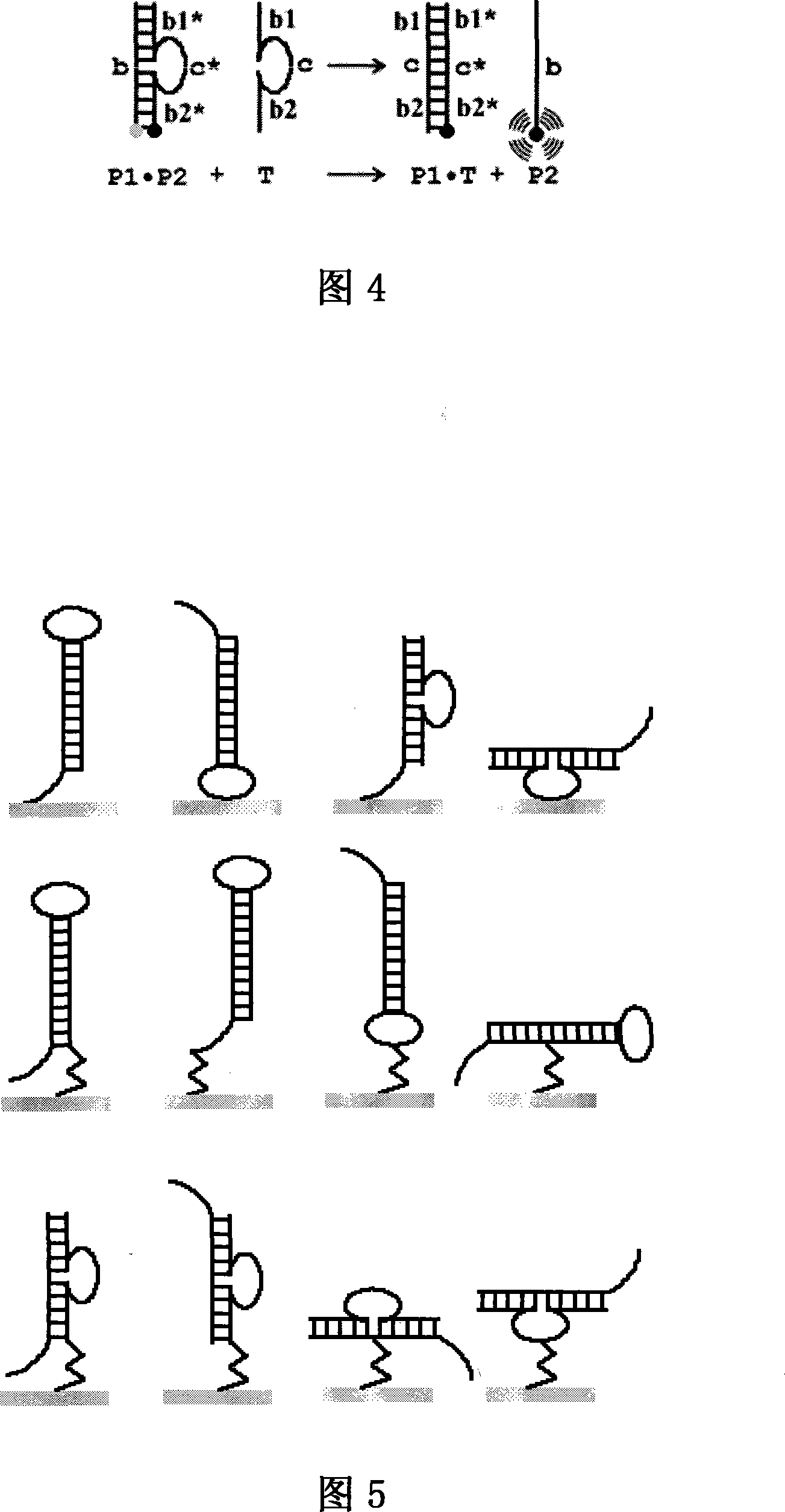
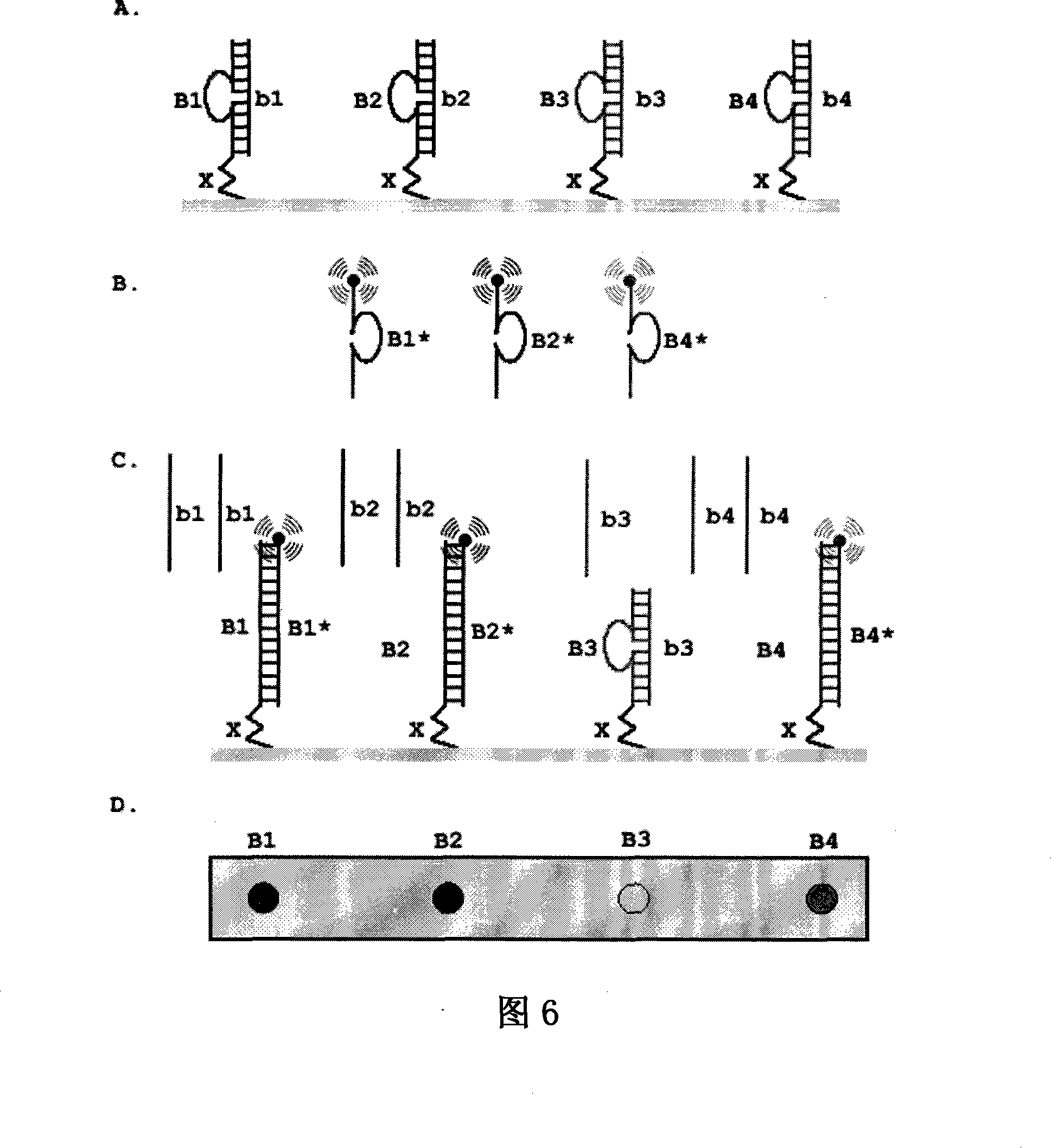
[0082] Example 3. Two-body probes with cohesive ends
[0083] A double oligonucleotide probe P1·P2 with a stem-loop structure, the probe is composed of two oligonucleotide chains complementary to each other, wherein the oligonucleotide chain P1 consists of the stem sequence b and The sticky end sequence a is connected; the oligonucleotide chain P2 is composed of the sequence b1*, c, b2*, wherein, b1* and b2* are complementary to the two ends of b of P1 respectively, and b1* and b2* are in the whole It is completely complementary to b, and c forms the ring part of the double-body probe; a fluorescent group is labeled at the end of P2, and a quencher group is labeled at the symmetry between the ends of P1 and P2 (that is, at the junction of b and a). The region consists of a stem b and a sticky end a. The target molecule T is formed by connecting the sequences b* and a*, and is completely complementary to the stem sequence b and the sticky end a of the probe P1·P2. When there ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com