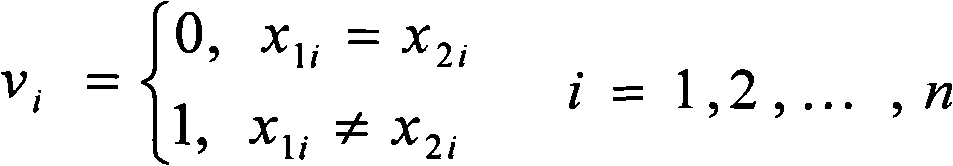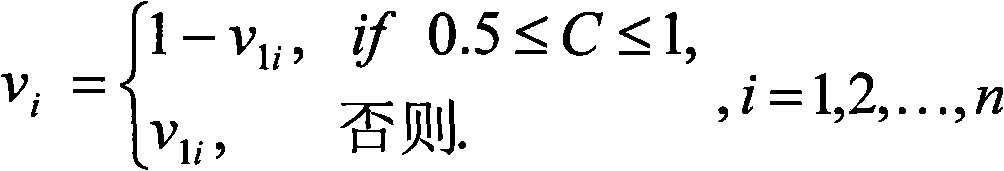Method for rebuilding individual single somatotype based on optimizing solution aggregate
A haplotype and optimized solution technology, applied in the field of bioinformatics, to achieve high haplotype reconstruction rate, high practical value, and reduce loss probability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
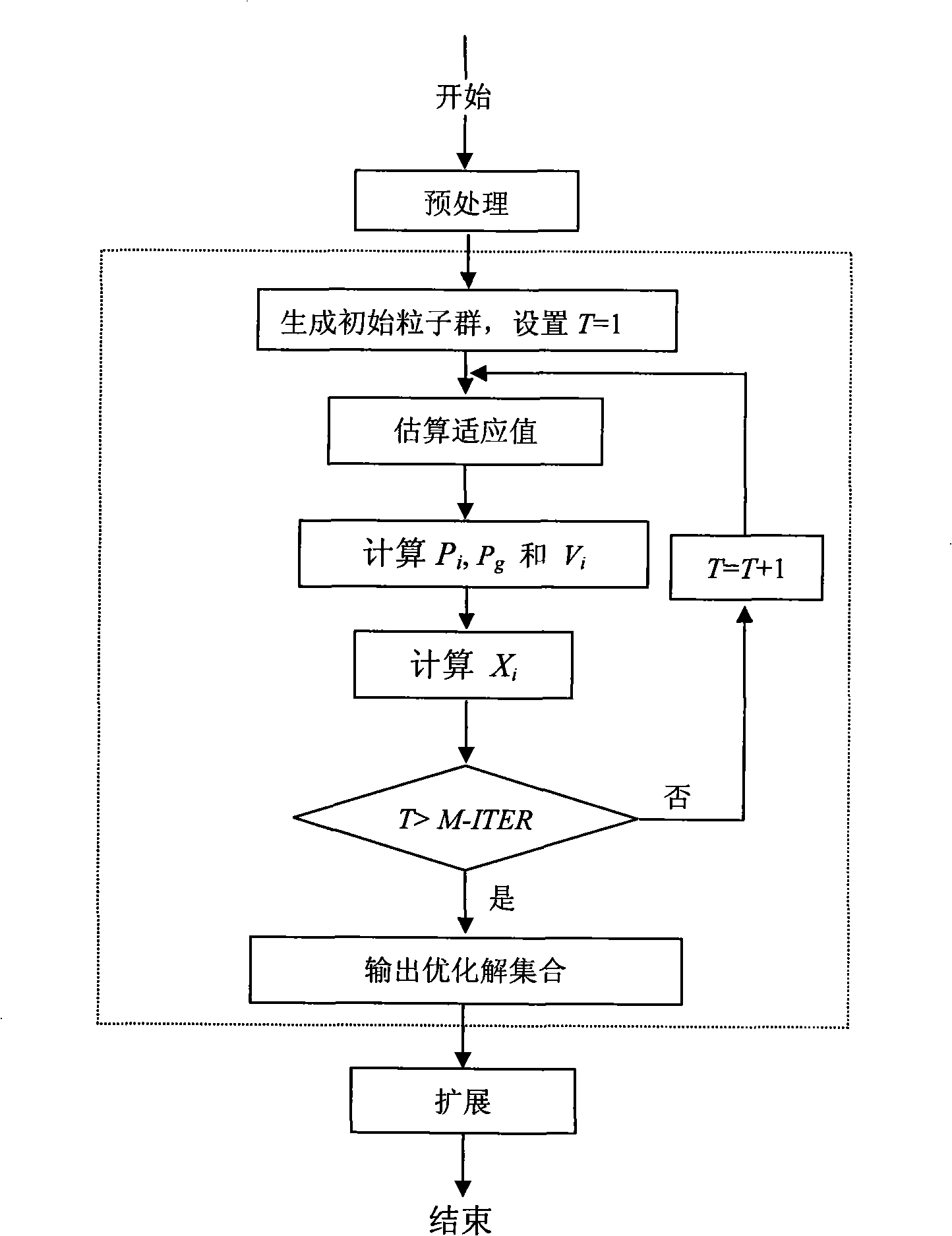
[0016] The specific implementation of the present invention will be further described below in conjunction with the accompanying drawings. see figure 1 , figure 1 It is a flow chart of the present invention, and the dotted box represents the particle swarm optimization method. Pretreatment SNP matrix M in the present invention m×n , to remove redundant information that is not helpful to the reconstruction work, that is, to delete all satisfying conditions f in M 0 ≤t or f 1 ≤t columns (in matrix M, let n x is the number of elements whose value is x in a certain column, and f x =n x / (n x +n 1-x )), where t is set to 0.2, if most of the non-empty elements in the deleted column are 0, it is called 0-column, otherwise it is called 1-column. After deleting all the columns that meet the above conditions, some rows will become empty rows (element values are all -), they are not helpful for the reconstruction work, so they are also deleted. After preprocessing, the SNP ma...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com